













中国农业科学 ›› 2021, Vol. 54 ›› Issue (2): 435-448.doi: 10.3864/j.issn.0578-1752.2021.02.018
陈华枝1( ),王杰1(
),王杰1( ),祝智威1,蒋海宾1,范元婵1,范小雪1,万洁琦1,卢家轩1,郑燕珍1,付中民1,2,3,徐国钧1,陈大福1,2,3,郭睿1,2,3(
),祝智威1,蒋海宾1,范元婵1,范小雪1,万洁琦1,卢家轩1,郑燕珍1,付中民1,2,3,徐国钧1,陈大福1,2,3,郭睿1,2,3( )
)
收稿日期:2020-02-10
接受日期:2020-02-25
出版日期:2021-01-16
发布日期:2021-02-03
通讯作者:
郭睿
作者简介:陈华枝,E-mail: 基金资助:
CHEN HuaZhi1( ),WANG Jie1(
),WANG Jie1( ),ZHU ZhiWei1,JIANG HaiBin1,FAN YuanChan1,FAN XiaoXue1,WAN JieQi1,LU JiaXuan1,ZHENG YanZhen1,FU ZhongMin1,2,3,XU GuoJun1,CHEN DaFu1,2,3,GUO Rui1,2,3(
),ZHU ZhiWei1,JIANG HaiBin1,FAN YuanChan1,FAN XiaoXue1,WAN JieQi1,LU JiaXuan1,ZHENG YanZhen1,FU ZhongMin1,2,3,XU GuoJun1,CHEN DaFu1,2,3,GUO Rui1,2,3( )
)
Received:2020-02-10
Accepted:2020-02-25
Online:2021-01-16
Published:2021-02-03
Contact:
Rui GUO
摘要:
【背景】蜜蜂球囊菌(Ascosphaera apis,简称球囊菌)是一种专性侵染蜜蜂幼虫的真菌病原,可引起成年蜜蜂数量和蜂群群势的急剧下降。长链非编码RNA(long non-coding RNA,lncRNA)是一类新近发现的非编码RNA,在表观遗传、细胞周期、剂量补偿等众多生命活动中发挥重要生物学功能。【目的】明确球囊菌菌丝和孢子中lncRNA的数量、种类和表达谱差异,并探究共有lncRNA、特有lncRNA和差异表达lncRNA(differentially expressed lncRNA,DElncRNA)在菌丝与孢子中的潜在功能。【方法】利用基于链特异性建库的lncRNA-seq技术对球囊菌的纯化菌丝(AaM)和纯化孢子(AaS)分别进行测序。根据FPKM(Fragment Per Kilobase of per Million mapped reads)法计算lncRNA在AaM和AaS中的表达水平。通过Venn分析筛选AaM与AaS的共有lncRNA和特有lncRNA。按照P≤0.05且|log2 fold change|≥1的标准筛选AaM vs AaS比较组中的DElncRNA。通过Blast工具将共有lncRNA、特有lncRNA和DElncRNA的上下游基因比对GO和KEGG数据库,以进行功能及通路注释。根据靶向结合关系构建共有lncRNA、特有lncRNA和DElncRNA的竞争性内源RNA(competing endogenous RNA,ceRNA)调控网络并利用Cytoscape软件进行可视化。利用RT-qPCR验证测序数据的可靠性。【结果】AaM和AaS分别测得108 614 646和105 675 408条原始读段(raw reads),经严格过滤得到107 780 032和104 621 402条有效读段(clean reads),Q20分别为98.76%和98.72%,Q30分别达到95.84%和95.78%。共鉴定到850个lncRNA。AaM和AaS的共有lncRNA为701个,二者的特有lncRNA分别为39和110个。上述共有lncRNA通过顺式作用调控3 992个上下游基因,它们涉及细胞进程、代谢进程和催化活性等42个功能条目,以及代谢途径、次生代谢物的生物合成和抗生素的生物合成等117条通路;AaM的特有lncRNA和AaS的特有lncRNA通过顺式作用分别调控243和672个上下游基因。AaM vs AaS比较组包含的255个DElncRNA通过顺式作用调控1 479个上下游基因,它们涉及代谢进程、细胞进程和催化活性等41个功能条目,以及代谢途径、次生代谢产物的生物合成和抗生素的生物合成等107条通路。从共有lncRNA、孢子特有lncRNA和DElncRNA中分别预测到41、5和13个微小RNA(microRNA,miRNA)的前体序列。调控网络分析结果显示,菌丝lncRNA、孢子lncRNA形成较为复杂的ceRNA调控网络;菌丝lncRNA可靶向结合8个miRNA,进而调控77个mRNA;孢子lncRNA可靶向结合7个miRNA,进而调控87个mRNA;2个DElncRNA(TCONS_00008630与TCONS_00009302)可靶向结合miR-4968-y,进而调控10个mRNA。RT-qPCR验证结果显示4个DElncRNA的差异表达趋势与测序结果一致,表明测序数据真实可靠。【结论】共有lncRNA、特有lncRNA和DElncRNA可能通过调控上下游基因的表达,作为miRNA的前体,以及充当ceRNA影响菌丝和孢子的物质和能量代谢、自噬、转录、MAPK信号通路、泛素介导的蛋白水解、蛋白酶体以及次生代谢产物的生物合成等生物学过程,从而调节球囊菌的生长、发育、生殖和致病性。
陈华枝,王杰,祝智威,蒋海宾,范元婵,范小雪,万洁琦,卢家轩,郑燕珍,付中民,徐国钧,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中长链非编码RNA的比较及潜在功能分析[J]. 中国农业科学, 2021, 54(2): 435-448.
CHEN HuaZhi,WANG Jie,ZHU ZhiWei,JIANG HaiBin,FAN YuanChan,FAN XiaoXue,WAN JieQi,LU JiaXuan,ZHENG YanZhen,FU ZhongMin,XU GuoJun,CHEN DaFu,GUO Rui. Comparison and Potential Functional Analysis of Long Non-Coding RNAs Between Ascosphaera apis Mycelium and Spore[J]. Scientia Agricultura Sinica, 2021, 54(2): 435-448.
表1
本研究所用的引物"
| LncRNA名称 LncRNA ID | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|
| TCONS_00006988 | F | CTCCAGTTGTGCGTTCAT |
| R | GTTGTCACCGTCTCTTCC | |
| TCONS_00003707 | F | GGAATGAATGATGCCAACTT |
| R | GTAGACCGAGGAAGAACAG | |
| TCONS_00001814 | F | ACAAGGAGGAAGTCAAGGA |
| R | CGAGCATAAGCAGTAGAGAT | |
| TCONS_00007359 | F | CCATCGCCACGGATATTC |
| R | CCAGAGCACATCAACATCA | |
| actin | F | CAGGAAAGGCTATGTTCGC |
| R | ATTACCGAGGAGCAAGACG |
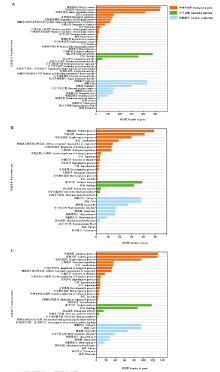
图1
共有和特有lncRNA的上下游基因的GO数据库注释 A:AaM与AaS共有lncRNA上下游基因的GO数据库注释GO database annotation of upstream and downstream genes of common lncRNAs in AaM and AaS;B:AaM特有lncRNA的GO数据库注释GO database annotation of upstream and downstream genes of specific lncRNAs in AaM;C:AaS特有lncRNA的GO数据库注释GO database annotation of upstream and downstream genes of specific lncRNAs in AaS"

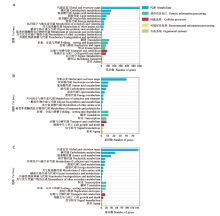
图2
共有和特有lncRNA上下游基因的KEGG富集分析 A:AaM与AaS共有lncRNA上下游基因的KEGG富集分析KEGG database annotation of upstream and downstream genes of common lncRNAs in AaM and AaS;B:AaM特有lncRNA的KEGG富集分析KEGG database annotation of upstream and downstream genes of specific lncRNAs in AaM;C:AaS特有lncRNA的KEGG富集分析KEGG database annotation of upstream and downstream genes of specific lncRNAs in AaS"
| [1] | CALDERÓN R A, RIVERA G, SÁNCHEZ L A, ZAMORA L G. Chalkbrood (Ascosphaera apis) and some other fungi associated with Africanized honey bees (Apis mellifera) in Costa Rica. Journal of Apicultural Research, 2004,43(4):187-188. |
| [2] | WOOD M. Microbes help bees battle chalkbrood. Agricultural Research, 1998,46(8):16-17. |
| [3] | ARONSTEINK K A, MURRAY D. Chalkbrood disease in honey bees. Journal of Invertebrate Pathology, 2010,103(Suppl.1):20-29. |
| [4] |
KAPRANOV P, CHENG J, DIKE S, NIX D A, DUTTAGUPTA R, WILLINGHAM A T, STADLER P F, HERTEL J, HACKERMÜLLER J, HOFACKER I L, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 2007,316(5830):1484-1488.
pmid: 17510325 |
| [5] |
PONTIER D B, GRIBNAU J. Xist regulation and function explored. Human Genetics, 2011,130(2):223-236.
pmid: 21626138 |
| [6] |
KINO T, HURT D E, ICHIJO T, NADER N, CHROUSOS G P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science Signaling, 2010, 3(107): ra8.
pmid: 20124551 |
| [7] | HEO J B, SUNG S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science, 2011,331(6013):76-79. |
| [8] | TSAI M C, MANOR O, WAN Y, MOSAMMAPARAST N, WANG J K, LAN F, SHI Y, SEGAL E, CHANG H Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science, 2010,329(5992):689-693. |
| [9] | MERCER T R, MATTICK J S. Structure and function of long noncoding RNAs in epigenetic regulation. Nature Structural and Molecular Biology, 2013,20(3):300-307. |
| [10] | LI M, SUN X, CAI H, SUN Y, PLATH M, LI C, LAN X, LEI C, LIN F, BAI Y, CHEN H. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochimica et Biophysica Acta, 2016,1859(7):871-882. |
| [11] |
ST LAURENT G, WAHLESTEDT C, KAPRANOV P. The landscape of long noncoding RNA classification. Trends in Genetics, 2015,31(5):239-251.
pmid: 25869999 |
| [12] | LI Z X, HAN K W, ZHANG D F, CHEN J G, XU Z, HOU L J. The role of long noncoding RNA in traumatic brain injury. Neuropsychiatric Disease and Treatment, 2019,15:1671-1677. |
| [13] | 郭睿, 王海朋, 陈华枝, 熊翠玲, 郑燕珍, 付中民, 赵红霞, 陈大福. 蜜蜂球囊菌的microRNA鉴定及其调控网络分析. 微生物学报, 2018,58(6):1077-1089. |
| GUO R, WANG H P, CHEN H Z, XIONG C L, ZHENG Y Z, FU Z M, ZHAO H X, CHEN D F. Identification of Ascosphaera apis microRNAs and investigation of their regulation networks. Acta Microbiologica Sinica, 2018,58(6):1077-1089. (in Chinese) | |
| [14] | KOPP F, MENDELL J T. Functional classification and experimental dissection of long noncoding RNAs. Cell, 2018,172(3):393-407. |
| [15] | KIM W, MIGUEL-ROJAS C, WANG J, TOWNSEND J P, TRAIL F. Developmental dynamics of long noncoding RNA expression during sexual fruiting body formation in Fusarium graminearum. mBio, 2018,9(4):e01292-18. |
| [16] | HIRIART E, VERDEL A. Long noncoding RNA-based chromatin control of germ cell differentiation: a yeast perspective. Chromosome Research, 2013,21(6/7):653-663. |
| [17] | CHEN D F, CHEN H Z, DU Y, ZHOU D D, GENG S H, WANG H P, WAN J Q, XIONG C L, ZHENG Y Z, GUO R. Genome-wide identification of long non-coding RNAs and their regulatory networks involved in Apis mellifera ligustica response to Nosema ceranae infection. Insects, 2019,10(8):245. |
| [18] |
WILUSZ J E, SUNWOO H, SPECTOR D L. Long noncoding RNAs: Functional surprises from the RNA world. Genes and Development, 2009,23(13):1494-1504.
doi: 10.1101/gad.1800909 pmid: 19571179 |
| [19] | FAN G Q, WANG Z, ZHAI X Q, CAO Y B. ceRNA cross-talk in paulownia witches’ broom disease. International Journal of Molecular Sciences, 2018,19(8):2463. |
| [20] |
QIN X, EVANS J D, ARONSTEIN K A, MURRAY K D, WEINSTOCK G M. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Molecular Biology, 2006,15(5):715-718.
pmid: 17069642 |
| [21] | SPILTOIR C F. Life cycle of Ascosphaera apis (Pericystis apis). American Journal of Botany, 1955,42(6):501-508. |
| [22] | FLORES J M, SPIVAK M, GUTIÉRREZ I. Spores of Ascosphaera apis contained in wax foundation can infect honeybee brood. Veterinary Microbiology, 2005,108(1/2):141-144. |
| [23] | SKOU J P. More details in support of the class Ascosphaeromycetes. Mycotaxon, 1988,31(1):191-198. |
| [24] | ANDERSON D, GIACON H SEARCH ARTICLES BY 'GIACON H' GIACON H, GIBSON N SEARCH ARTICLES BY 'GIBSON N' GIBSON N. Detection and thermal destruction of the chalkbrood fungus (Ascosphaera apis) in honey. Journal of Apicultural Research, 1997,36(3/4):163-168. |
| [25] |
SHANG Y F, XIAO G H, ZHENG P, CEN K, ZHAN S, WANG C S. Divergent and convergent evolution of fungal pathogenicity. Genome Biology and Evolution, 2016,8(5):1374-1387.
pmid: 27071652 |
| [26] | 郭睿, 耿四海, 熊翠玲, 郑燕珍, 付中民, 王海朋, 杜宇, 童新宇, 赵红霞, 陈大福. 意大利蜜蜂工蜂中肠发育过程中长链非编码RNA的差异表达分析. 中国农业科学, 2018,51(18):3600-3613. |
| GUO R, GENG S H, XIONG C L, ZHENG Y Z, FU Z M, WANG H P, DU Y, TONG X Y, ZHAO H X, CHEN D F. Differential expression analysis of long non-coding RNAs during the developmental process of Apis mellifera ligustica worker’s midgut. Scientia Agricultura Sinica, 2018,51(18):3600-3613. (in Chinese) | |
| [27] | FLÓREZ-ZAPATA N M V, REYES-VALDÉS M H, MARTÍNEZ O. Long non-coding RNAs are major contributors to transcriptome changes in sunflower meiocytes with different recombination rates. BMC Genomics, 2016,17:490. |
| [28] | YANG L, LONG Y P, LI C, CAO L, GAN H Y, HUANG K L, JIA Y J. Genome-wide analysis of long noncoding RNA profile in human gastric epithelial cell response to Helicobacter pylori. Japanese Journal of Infectious Diseases, 2015,68(1):63-66. |
| [29] | GUO R, CHEN D F, XIONG C L, HOU C S, ZHENG Y Z, FU Z M, LIANG Q, DIAO Q Y, ZHANG L, WANG H Q, HOU Z X, KUMAR D. First identification of long non-coding RNAs in fungal parasite Nosema ceranae. Apidologie, 2018,49(5):660-670. |
| [30] | WANG Z Q, ZHAO Y W, ZHANG Y. Viral lncRNA: A regulatory molecule for controlling virus life cycle. Noncoding RNA Research, 2017,2(1):38-44. |
| [31] |
GUO R, CHEN D F, XIONG C L, HOU C S, ZHENG Y Z, FU Z M, DIAO Q Y, ZHANG L, WANG H Q, HOU Z X, LI W D, DHIRAJ K, LIANG Q. Identification of long non-coding RNAs in the chalkbrood disease pathogen Ascospheara apis. Journal of Invertebrate Pathology, 2018,156:1-5.
pmid: 29894727 |
| [32] | GUO R, CHEN D F, CHEN H Z, FU Z M, XIONG C L, HOU C S, ZHENG Y Z, GUO Y, WANG H P, DU Y, DIAO Q Y. Systematic investigation of circular RNAs in Ascosphaera apis, a fungal pathogen of honeybee larvae. Gene, 2018,678:17-22. |
| [33] | 陈大福, 郭睿, 熊翠玲, 梁勤, 郑燕珍, 徐细建, 黄枳腱, 张曌楠, 张璐, 李汶东, 童新宇, 席伟军. 胁迫意大利蜜蜂幼虫肠道的球囊菌的转录组分析. 昆虫学报, 2017,60(4):401-411. |
| CHEN D F, GUO R, XIONG C L, LIANG Q, ZHENG Y Z, XU X J, HUANG Z J, ZHANG Z N, ZHANG L, LI W D, TONG X Y, XI W J. Transcriptomic analysis of Ascosphaera apis stressing larval gut of Apis mellifera ligustica (Hyemenoptera: Apidae). Acta Entomologica Sinica, 2017,60(4):401-411. (in Chinese) | |
| [34] | 张曌楠, 熊翠玲, 徐细建, 黄枳腱, 郑燕珍, 骆群, 刘敏, 李汶东, 童新宇, 张琦, 梁勤, 郭睿, 陈大福. 蜜蜂球囊菌的参考转录组de novo组装及SSR分子标记开发. 昆虫学报, 2017,60(1):34-44. |
| ZHANG Z N, XIONG C L, XU X J, HUANG Z J, ZHENG Y Z, LUO Q, LIU M, LI W D, TONG X Y, ZHANG Q, LIANG Q, GUO R, CHEN D F. De novo assembly of a reference transcriptome and development of SSR markers for Ascosphaera apis. Acta Entomologica Sinica, 2017,60(1):34-44. (in Chinese) | |
| [35] | 陈大福, 郭睿, 熊翠玲, 梁勤, 郑燕珍, 徐细建, 张曌楠, 黄枳腱, 张璐, 王鸿权, 解彦玲, 童新宇. 中华蜜蜂幼虫肠道响应球囊菌早期胁迫的转录组学. 中国农业科学, 2017,50(13):2614-2623. |
| CHEN D F, GUO R, XIONG C L, LIANG Q, ZHENG Y Z, XU X J, ZHANG Z N, HUANG Z J, ZHANG L, WANG H Q, XIE Y N, TONG X Y. Transcriptome of Apis cerana cerana larval gut under the stress of Ascosphaera apis. Scientia Agricultura Sinica, 2017,50(13):2614-2623. (in Chinese) | |
| [36] | LANGMEAD B, TRAPNELL C, POP M, SALZBERG S L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 2009,10(3):R25. |
| [37] |
KIM D, PERTEA G, TRAPNELL C, PIMENTEL H, KELLEY R, SALZBERG S L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 2013,14(4):R36.
pmid: 23618408 |
| [38] |
TRAPNELL C, ROBERTS A, GOFF L, PERTEA G, KIM D, KELLEY D R, PIMENTEL H, SALZBERG S L, RINN J L, PACHTER L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 2012,7(3):562-578.
doi: 10.1038/nprot.2012.016 pmid: 22383036 |
| [39] | KONG L, ZHANG Y, YE Z Q, LIU X Q, ZHAO S Q, WEI L P, GAO G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research, 2007,35:W345-349. |
| [40] |
SUN L, LUO H T, BU D C, ZHAO G G, YU K T, ZHANG C H, LIU Y N, CHEN R S, ZHAO Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research, 2013,41(17):e166.
doi: 10.1093/nar/gkt646 pmid: 23892401 |
| [41] |
ROBINSON M D, MCCARTHY D J, SMYTH G K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010,26(1):139-140.
pmid: 19910308 |
| [42] |
SHAO J, CHEN H, YANG D, JIANG M, ZHANG H, WU B, LI J, YUAN L, LIU C. Genome-wide identification and characterization of natural antisense transcripts by strand-specific RNA sequencing in Ganoderma lucidum. Scientific Reports, 2017,7(1):5711.
pmid: 28720793 |
| [43] | 陈华枝, 祝智威, 蒋海宾, 王杰, 范元婵, 范小雪, 万洁琦, 卢家轩, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 蜜蜂球囊菌菌丝和孢子中微小RNA及其靶mRNA的比较分析. 中国农业科学, 2020,53(17):3606-3619. |
| CHEN H Z, ZHU Z W, JIANG H B, WANG J, FAN Y C, FAN X X, WAN J Q, LU J X, XIONG C L, ZHENG Y Z, FU Z M, CHEN D F, GUO R. Comparative analysis of microRNAs and corresponding target mRNAs in Ascosphaera apis mycelium and spore. Scientia Agricultura Sinica, 2020,53(17):3606-3619. (in Chinese) | |
| [44] | 熊翠玲, 陈华枝, 陈大福, 郑燕珍, 付中民, 徐国钧, 杜宇, 王海朋, 耿四海, 周丁丁, 刘思亚, 郭睿. 意大利蜜蜂工蜂中肠的环状RNA及其调控网络分析. 昆虫学报, 2018,61(12):1363-1375. |
| XIONG C L, CHEN H Z, CHEN D F, ZHENG Y Z, FU Z M, XU G J, DU Y, WANG H P, GENG S H, ZHOU D D, LIU S Y, GUO R. Analysis of circular RNAs and their regulatory networks in the midgut of Apis mellifera ligustica workers. Acta Entomologica Sinica, 2018,61(12):1363-1375. (in Chinese) | |
| [45] |
ALLEN E, XIE Z, GUSTAFSON A M, CARRINGTON J C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 2005,121(2):207-221.
doi: 10.1016/j.cell.2005.04.004 pmid: 15851028 |
| [46] |
ATIANAND M K, FITZGERALD K A. Long non-coding RNAs and control of gene expression in the immune system. Trends in Molecular Medicine, 2014,20(11):623-631.
pmid: 25262537 |
| [47] |
QURESHI I A, MEHLER M F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature Reviews. Neuroscience, 2012,13(8):528-541.
doi: 10.1038/nrn3234 pmid: 22814587 |
| [48] |
PAULI A, VALEN E, LIN M F, GARBER M, VASTENHOUW N L, LEVIN J Z, FAN L, SANDELIN A, RINN J L, REGEV A, SCHIER A F. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Research, 2012,22(3):577-591.
doi: 10.1101/gr.133009.111 pmid: 22110045 |
| [49] |
WANG Z, JIANG Y, WU H, XIE X, HUANG B. Genome-wide identification and functional prediction of long non-coding RNAs involved in the heat stress response in Metarhizium robertsii. Frontiers in Microbiology, 2019,10:2336.
doi: 10.3389/fmicb.2019.02336 pmid: 31649657 |
| [50] | YORIMITSU T, KLIONSKY D J. Autophagy: Molecular machinery for self-eating. Cell Death and Differentiation, 2005,12(Suppl. 2):1542-1552. |
| [51] | FAN C L, CHANG A N, LIU T B. Role of autophagy in the reproduction of pathogenic fungi. Acta Microbiologica Sinica, 2019,59(2):224-234. |
| [52] |
LIU X H, ZHAO Y H, ZHU X M, ZENG X Q, HUANG L Y, DONG B, SU Z Z, WANG Y, LU J P, LIN F C. Autophagy-related protein MoAtg14 is involved in differentiation, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Scientific Reports, 2017,7:40018.
pmid: 28067330 |
| [53] |
DENG Y Z, RAMOS-PAMPLONA M, NAQVI N I. Autophagy- ssisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy, 2009,5(1):33-43.
doi: 10.4161/auto.5.1.7175 pmid: 19115483 |
| [54] |
WANG P, GRANADOS R R. Observations on the presence of the peritrophic membrane in larval Trichoplusia ni and its role in limiting baculovirus infection. Journal of Invertebrate Pathology, 1998,72(1):57-62.
doi: 10.1006/jipa.1998.4759 pmid: 9647702 |
| [55] |
SURESH B, LEE J, KIM K S, RAMAKRISHNA S. The importance of ubiquitination and deubiquitination in cellular reprogramming. Stem Cells International, 2016,2016:6705927.
doi: 10.1155/2016/6705927 pmid: 26880980 |
| [56] |
BIDOCHKA M J, KHACHATOURIANS G G. The implication of metabolic acids produced by Beauveria bassiana in pathogenesis of the migratory grasshopper, Melanoplus sanguinipes. Journal of Invertebrate Pathology, 1991,58(1):106-117.
doi: 10.1016/0022-2011(91)90168-P |
| [57] | GOTZ P, MATHA V, VILCINSKAS A. Effects of the entomopathogenic fungus Metarhizium anisopliae and its secondary metabolites on morphology and cytoskeleton of plasmatocytes isolated from the greater wax moth, Galleria mellonella. Journal of Invertebrate Pathology, 1997,43(12):1149-1159. |
| [58] |
JENSEN A B, ARONSTEIN K, FLORES J M, VOJVODIC S, PALACIO M A, SPIVAK M. Standard methods for fungal brood disease research. Journal of Apicultural Research, 2013, 52(1): 10.3896/IBRA.1.52.1.13.
doi: 10.3896/IBRA.1.52.1.13 pmid: 24198438 |
| [59] | BROWN A J P, BUDGE S, KALORITI D, TILLMANN A, JACOBSEN M D, YIN Z, ENE I V, BOHOVYCH I, SANDAI D, KASTORA S, POTRYKUS J, BALLOU E R, CHILDERS D S, SHAHANA S, LEACH M D. Stress adaptation in a pathogenic fungus. Journal of Experimental Biology, 2014,217(1):144-155. |
| [60] |
SO K K, KIM D H. Role of MAPK signaling pathways in regulating the hydrophobin cryparin in the chestnut blight fungus Cryphonectria parasitica. Mycobiology, 2017,45(4):362-369.
doi: 10.5941/MYCO.2017.45.4.362 pmid: 29371804 |
| [61] |
JIANG C, ZHANG X, LIU H Q, XU J R. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathogens, 2018,14(3):e1006875.
pmid: 29543901 |
| [62] | NI X, GAO J X, YU C J, WANG M, SUN J N, LI Y Q, CHEN J. MAPKs and acetyl-CoA are associated with Curvularia lunata pathogenicity and toxin production in maize. Journal of Integrative Agriculture, 2018,17(1):139-148. |
| [63] |
SALMENA L, POLISENO L, TAY Y, KATS L, PANDOLFI P P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell, 2011,146(3):353-358.
pmid: 21802130 |
| [64] |
HUANG M J, ZHAO J Y, XU J J, LI Y, ZHUANG Y F, ZHANG X L. LncRNA ADAMTS9-AS2 controls human mesenchymal stem cell chondrogenic differentiation and functions as a ceRNA. Molecular Therapy-Nucleic Acids, 2019,18:533-545.
pmid: 31671346 |
| [1] | 李晓菁,张思雨,刘迪,袁晓伟,李兴盛,石延霞,谢学文,李磊,范腾飞,李宝聚,柴阿丽. 芸薹根肿菌活细胞PMAxx-qPCR快速定量检测方法的建立与应用[J]. 中国农业科学, 2022, 55(10): 1938-1948. |
| [2] | 杜宇,范小雪,蒋海宾,王杰,冯睿蓉,张文德,余岢骏,隆琦,蔡宗兵,熊翠玲,郑燕珍,陈大福,付中民,徐国钧,郭睿. 微小RNA介导意大利蜜蜂工蜂对东方蜜蜂微孢子虫的跨界调控[J]. 中国农业科学, 2021, 54(8): 1805-1820. |
| [3] | 陈华枝,范元婵,蒋海宾,王杰,范小雪,祝智威,隆琦,蔡宗兵,郑燕珍,付中民,徐国钧,陈大福,郭睿. 基于纳米孔全长转录组数据完善东方蜜蜂微孢子虫的基因组注释[J]. 中国农业科学, 2021, 54(6): 1288-1300. |
| [4] | 杜宇,祝智威,王杰,王秀娜,蒋海宾,范元婵,范小雪,陈华枝,隆琦,蔡宗兵,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 利用第三代纳米孔长读段测序技术构建和注释蜜蜂球囊菌的全长转录组[J]. 中国农业科学, 2021, 54(4): 864-876. |
| [5] | 王继卿,郝志云,沈继源,柯娜,黄兆春,梁维炜,罗玉柱,胡江,刘秀,李少斌. 小尾寒羊泌乳性状重要lncRNAs的筛选、鉴定及功能分析[J]. 中国农业科学, 2021, 54(14): 3113-3123. |
| [6] | 周丁丁, 范元婵, 王杰, 蒋海宾, 祝智威, 范小雪, 陈华枝, 杜宇, 周紫彧, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 蜜蜂球囊菌中长链非编码RNA的调控作用[J]. 中国农业科学, 2021, 54(1): 224-238. |
| [7] | 贾海燕,宋丽云,徐翔,解屹,张超群,刘天波,赵存孝,申莉莉,王杰,李莹,王凤龙,杨金广. 不同温度下TMV侵染枯斑三生烟的LncRNA差异表达[J]. 中国农业科学, 2020, 53(7): 1381-1396. |
| [8] | 段应策,胡姿仪,杨帆,李金涛,邬向丽,张瑞颖. pH和缓冲作用对香菇菌丝生长的影响[J]. 中国农业科学, 2020, 53(22): 4683-4690. |
| [9] | 陈华枝,祝智威,蒋海宾,王杰,范元婵,范小雪,万洁琦,卢家轩,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中微小RNA及其靶mRNA的比较分析[J]. 中国农业科学, 2020, 53(17): 3606-3619. |
| [10] | 耿四海,石彩云,范小雪,王杰,祝智威,蒋海宾,范元婵,陈华枝,杜宇,王心蕊,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 微小RNA介导东方蜜蜂微孢子虫侵染意大利蜜蜂工蜂的分子机制[J]. 中国农业科学, 2020, 53(15): 3187-3204. |
| [11] | 杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿. 微小RNA及其介导的竞争性内源RNA调控网络在意大利蜜蜂工蜂中肠发育过程中的潜在作用[J]. 中国农业科学, 2020, 53(12): 2512-2526. |
| [12] | 周丁丁,史小玉,王杰,范元婵,祝智威,蒋海宾,范小雪,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 东方蜜蜂微孢子虫孢子中长链非编码RNA的竞争性内源RNA调控网络及潜在功能[J]. 中国农业科学, 2020, 53(10): 2122-2136. |
| [13] | 杜宇,周丁丁,万洁琦,卢家轩,范小雪,范元婵,陈恒,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 意大利蜜蜂工蜂中肠发育过程中的差异基因 表达谱及调控网络[J]. 中国农业科学, 2020, 53(1): 201-212. |
| [14] | 付中民,陈华枝,刘思亚,祝智威,范小雪,范元婵,万洁琦,张璐,熊翠玲,徐国钧,陈大福,郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答[J]. 中国农业科学, 2019, 52(17): 3069-3082. |
| [15] | 易敏,吕青,刘柯柯,王礼君,吴玉娇,周泽扬,龙梦娴. 家蚕微孢子虫极管蛋白2(NbPTP2)的表达、纯化和定位特征[J]. 中国农业科学, 2019, 52(10): 1830-1838. |
|
||