













中国农业科学 ›› 2020, Vol. 53 ›› Issue (10): 2122-2136.doi: 10.3864/j.issn.0578-1752.2020.10.018
• 畜牧·兽医·资源昆虫 • 上一篇
周丁丁,史小玉,王杰,范元婵,祝智威,蒋海宾,范小雪,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿( )
)
收稿日期:2019-10-23
接受日期:2019-11-26
出版日期:2020-05-16
发布日期:2020-05-22
通讯作者:
郭睿
作者简介:周丁丁,E-mail:zdd03569981@163.com。|史小玉,E-mail:2395965008@qq.com。
基金资助:
ZHOU DingDing,SHI XiaoYu,WANG Jie,FAN YuanChan,ZHU ZhiWei,JIANG HaiBin,FAN XiaoXue,XIONG CuiLing,ZHENG YanZhen,FU ZhongMin,XU GuoJun,CHEN DaFu,GUO Rui( )
)
Received:2019-10-23
Accepted:2019-11-26
Online:2020-05-16
Published:2020-05-22
Contact:
Rui GUO
摘要:
【背景】 长链非编码RNA(long non-coding RNA,lncRNA)是一类长度>200 nt且不具有编码能力的转录本,能够在真核生物的转录水平和转录后水平通过顺式(cis)、反式(trans)或竞争性内源RNA(competing endogenous RNA,ceRNA)机制调控基因的表达,已被证明在生物体的生长和发育等生物学过程中发挥重要的调控作用。【目的】 结合测序得到的东方蜜蜂微孢子虫(Nosema ceranae)纯化孢子的small RNA(sRNA)组学数据和前期获得的lncRNA组学数据对lncRNA的调控方式进行深入细致的分析和探讨,揭示lncRNA在东方蜜蜂微孢子虫孢子中的潜在功能。【方法】 利用small RNA-seq(sRNA-seq)技术对东方蜜蜂微孢子虫纯化孢子进行测序,通过相关的生物信息学软件对测序数据进行质控。根据lncRNA与mRNA的位置关系预测lncRNA的上下游基因;使用Blast软件比对GO和KEGG数据库,对上下游基因进行功能和代谢通路注释;利用Target Finder软件预测lncRNA-miRNA及lncRNA-miRNA-mRNA调控网络,进而通过Cytoscape v3.7.2软件对调控网络进行可视化;利用RT-PCR对调控网络中的部分lncRNA、miRNA和mRNA的表达进行验证。【结果】 东方蜜蜂微孢子虫纯化孢子样品的sRNA-seq分别得到16 597 883、15 451 791和12 248 316条raw reads,经质控分别得到15 608 370、14 249 255和11 440 684条clean reads,各样品的Q30≥98.58%。共预测出lncRNA的上下游基因310个,它们可注释到代谢进程、细胞进程、催化活性、结合和细胞等相关的35个功能条目;此外还可注释到56条代谢通路,涉及嘌呤代谢、碳代谢和丙酮酸代谢等物质代谢通路,以及甲烷代谢、氧化磷酸化和糖酵解/糖异生等能量代谢通路。上述结果表明相应的lncRNA可能通过cis作用调节上下游基因的表达,从而参与调控东方蜜蜂微孢子虫孢子中的物质和能量代谢以及细胞生命活动。进一步构建lncRNA-miRNA调控网络,分析结果显示MSTRG.3636.1、MSTRG.4498.1和MSTRG.4883.1可靶向结合4个miRNA(nce-miR-7729、nce-miR-7502、nce-miR-8639和nce-miR-8565),说明此3个lncRNA可作为ceRNA在东方蜜蜂微孢子虫孢子中发挥潜在作用。LncRNA-miRNA-mRNA调控网络分析结果显示,三者之间形成较为复杂的调控网络关系,miRNA处于调控网络的中心并联系lncRNA和mRNA,nce-miR-7502和nce-miR-8639结合的mRNA最多,达到28个;与MSTRG.3636.1、MSTRG.4498.1和MSTRG.4883.1存在靶向关系的miRNA结合多个mRNA,表明上述3个lncRNA可能通过ceRNA机制发挥作用,从而影响东方蜜蜂微孢子虫孢子中的新陈代谢和生命活动。【结论】 lncRNA可能通过cis作用调控上下游基因的表达,以及作为ceRNA通过吸附miRNA间接影响靶基因的表达,从而参与调节东方蜜蜂微孢子虫孢子中的物质和能量代谢等基本生命活动。
周丁丁,史小玉,王杰,范元婵,祝智威,蒋海宾,范小雪,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 东方蜜蜂微孢子虫孢子中长链非编码RNA的竞争性内源RNA调控网络及潜在功能[J]. 中国农业科学, 2020, 53(10): 2122-2136.
ZHOU DingDing,SHI XiaoYu,WANG Jie,FAN YuanChan,ZHU ZhiWei,JIANG HaiBin,FAN XiaoXue,XIONG CuiLing,ZHENG YanZhen,FU ZhongMin,XU GuoJun,CHEN DaFu,GUO Rui. Investigation of Competing Endogenous RNA Regulatory Network and Putative Function of Long Non-Coding RNAs in Nosema ceranae Spore[J]. Scientia Agricultura Sinica, 2020, 53(10): 2122-2136.
表1
本研究使用的引物"
| 核酸ID Nucleotide ID | 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 预期目的片段 Product size (bp) |
|---|---|---|---|
| gene1439 | 1-F | ATGTATCTTATGCCAACGC | 382 |
| 1-R | TGAATCTTTCCTACACCCT | ||
| gene565 | 2-F | TCAAGCCTGTTAAGATGAC | 499 |
| 2-R | ACTACTTCTGGCGGTTTAT | ||
| gene1719 | 3-F | ATAGGAGGTAAACATCAG | 438 |
| 3-R | ATAGTAAAGTTCAGTGCC | ||
| gene1366 | 4-F | TTGAAGAAGGTGTCGCAGAAC | 182 |
| 4-R | AGCAGAGCCACATAAACAAGC | ||
| gene1360 | 5-F | AAGAAGCCGAAACAACCC | 101 |
| 5-R | GTTAGCCGCATCCATAGC | ||
| gene1134 | 6-F | TGTACGGTCTGGCTGCTTC | 352 |
| 6-R | GGACTTGGCTTTCCACTAA | ||
| gene1695 | 7-F | GATACAGCCTCTTTGG | 138 |
| 7-R | GGAACAATAACCCTAAA | ||
| gene1563 | 8-F | TGCCTACAATCAACTCTAAT | 384 |
| 8-R | CACCCAATACTTCAAATAAC | ||
| MSTRG.3636.1 | 9-F | TAACTAGCCTACTCTATCCC | 257 |
| 9-R | TAAAAGCACTAACTACAACC | ||
| MSTRG.4883.1 | 10-F | CTGACAGACCATAGTCGGCATA | 128 |
| 10-R | TCTACAATGATGGGCACAGGA | ||
| nce-miR-8565 | 11-L | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCTAAAGA | 67 |
| 11-F | CGCGCCTTTAATTGTGAAACATG | ||
| nce-miR-7502 | 12-L | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACTTGTC | 66 |
| 12-F | CGCGCCTAAAAACAAAAATGTA | ||
| nce-miR-7729 | 13-L | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCTAGGCC | 62 |
| 13-F | CGCGCCTAGTGATGGCTGT | ||
| - | R | CTCAACTGGTGTCGTGGA |
表4
东方蜜蜂微孢子虫孢子中lncRNA靶向miRNA的靶mRNA的Swissprot和Nr数据库注释"
| 核酸ID Nucleotide ID | Swissprot 数据库注释 Swissprot database annotation | Nr数据库注释 Nr database annotation |
|---|---|---|
| gene986 | 液泡氨基酸转运体5 Vacuolar amino acid transporter 5 | 假定蛋白NCER_101697 Hypothetical protein NCER_101697 |
| gene783 | 负辅因子2复合亚基 Negative cofactor 2 complex subunit beta | 假定蛋白NCER_101896 Hypothetical protein NCER_101896 |
| gene565 | 丝氨酸/苏氨酸多聚蛋白激酶1 Serine/threonine-protein kinase plo1 | 假定蛋白NCER__102113 Hypothetical protein NCER_102113 |
| gene2596 | 含C1778.09蛋白的TBC结构域 TBC domain-containing protein C1778.09 | 假定蛋白NCER_100108 Hypothetical protein NCER_100108 |
| gene2543 | TATA结合蛋白相关因子MOT1 TATA-binding protein-associated factor MOT1 | 假定蛋白NCER_100152 Hypothetical protein NCER_100152 |
| gene2397 | 丝氨酸/苏氨酸蛋白激酶CDC5同源物 Cell cycle serine/threonine-protein kinase CDC5 homolog | 假定蛋白NCER_100270 Hypothetical protein NCER_100270 |
| gene2312 | ERAD-相关E3泛素蛋白连接酶doa10 ERAD-associated E3 ubiquitin-protein ligase doa10 | 假定蛋白NCER_100375 Hypothetical protein NCER_100375 |
| gene2178 | 转运蛋白sec24 Transport protein sec24 | 假定蛋白NCER_100510 Hypothetical protein NCER_100510 |
| gene2014 | 转录相关蛋白1 Transcription-associated protein 1 | 假定蛋白NCER_100665 Hypothetical protein NCER_100665 |
| gene1870 | ERAD-相关E3泛素蛋白连接酶doa10 ERAD-associated E3 ubiquitin-protein ligase doa10 | 假定蛋白NCER_100805 Hypothetical protein NCER_100805 |
| gene1766 | 赖氨酸- tRNA连接酶 Lysine-tRNA ligase | 假定蛋白NCER_100909 Hypothetical protein NCER_100909 |
| gene1719 | T-复合体蛋白1亚基 T-complex protein 1 subunit alpha | 假定蛋白NCER_100958 Hypothetical protein NCER_100958 |
| gene1687 | DNA-锚定 RNA 聚合酶 II 亚基RPB2 DNA-directed RNA polymerase II subunit RPB2 | 假定蛋白NCER_100983 Hypothetical protein NCER_100983 |
| gene1678 | 锰-ATP转运酶4 Manganese-transporting ATPase 4 | 假定蛋白NCER_101005 Hypothetical protein NCER_101005 |
| gene1677 | 3型蛋白酶体亚基 Probable proteasome subunit beta type-3 | 假定蛋白NCER_101004 Hypothetical protein NCER_101004 |
| gene1662 | 微管蛋白γ链 Tubulin gamma chain | 假定蛋白NCER_101024 Hypothetical protein NCER_101024 |
| gene1563 | 核输出蛋白BRL1 Nucleus export protein BRL1 | 假定蛋白NCER_101114 Hypothetical protein NCER_101114 |
| gene1383 | V型质子ATP酶催化亚基A V-type proton ATPase catalytic subunit A | 假定蛋白NCER_101294 Hypothetical protein NCER_101294 |
| gene1366 | 长链脂肪酸CoA链2 Long chain fatty acid CoA ligase 2 | 假定蛋白NCER_101315 Hypothetical protein NCER_101315 |
| gene1360 | 热休克蛋白hsp98 Heat shock protein hsp98 | 假定蛋白NCER_101322 Hypothetical protein NCER_101322 |
| gene1348 | 丝氨酸/苏氨酸蛋白激酶MEC1同源物 Serine/threonine-protein kinase MEC1 homolog | 假定蛋白NCER_101332 Hypothetical protein NCER_101332 |
| gene1190 | 泛素羧基末端水解酶5 Ubiquitin carboxyl-terminal hydrolase 5 | 假定蛋白NCER_101487 Hypothetical protein NCER_101487 |
| gene1180 | 苯丙氨酸-tRNA连接酶亚基 Phenylalanine-tRNA ligase beta subunit | 假定蛋白NCER_101490 Hypothetical protein NCER_101490 |
| gene1134 | 二磷酸异戊基丁二醇异构酶 Isopentenyl-diphosphate Delta-isomerase | 假定蛋白NCER_101544 Hypothetical protein NCER_101544 |
| newGene_142 | 60S核糖体蛋白L40(前体) 60S ribosomal protein L40 (precursor) | 假定蛋白NCER_102067 Hypothetical protein NCER_102067 |
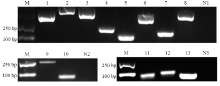
图5
东方蜜蜂微孢子虫孢子中lncRNA、靶miRNA和靶mRNA的RT-PCR验证 M:DNA marker;1:假定蛋白NCER_101243编码基因Hypothetical protein NCER_101243 coding gene;2:假定蛋白NCER_102113编码基因Hypothetical protein NCER_102113 coding gene;3:假定蛋白NCER_100958编码基因Hypothetical protein NCER_100958 coding gene;4:假定蛋白NCER_101315编码基因Hypothetical protein NCER_101315 coding gene;5:假定蛋白NCER_101322编码基因Hypothetical protein NCER_101322 coding gene;6:假定蛋白NCER_101544编码基因Hypothetical protein NCER_101544 coding gene;7:假定蛋白NCER_100991编码基因Hypothetical protein NCER_100991 coding gene;8:假定蛋白NCER_1011114编码基因Hypothetical protein NCER_101114 coding gene;9:MSTRG.3636.1;10:MSTRG.4883.1;11:nce-miR-8565;12:nce-miR-7502;13:nce-miR-7729;N1、N2:阴性对照(无菌水)Negative control (sterile water);N3:阴性对照(DEPC水)Negative control (DEPC water)"
| [1] | KEELING P J, FAST N M . Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annual Review of Microbiology, 2002,56:93-116. |
| [2] | HOLT H L, VILLAR G, CHENG W, SONG J, GROZINGER C M . Molecular, physiological and behavioral responses of honey bee ( Apis mellifera) drones to infection with microsporidian parasites. Journal of Invertebrate Pathology, 2018,155:14-24. |
| [3] | KURZE C, CONTE Y L, KRYGER P, LEWKOWSKI O, MÜLLER T, MORITZ R F A . Infection dynamics of Nosema ceranae in honey bee midgut and host cell apoptosis. Journal of Invertebrate Pathology, 2018,154:1-4. |
| [4] | ARNDT K M, REINES D . Termination of transcription of short noncoding RNAs by RNA polymerase II. Annual Review of Biochemistry, 2015,84:381-404. |
| [5] | WU Z M, LIU X X, LIU L, DENG H L, ZHANG J J, XU Q, CEN B H, JI A M . Regulation of lncRNA expression. Cellular and Molecular Biology Letters, 2014,19(4):561-575. |
| [6] | KOPP F, MENDELL J T . Functional classification and experimental dissection of long noncoding RNAs. Cell, 2018,172(3):393-407. |
| [7] | DE BOER P, VOS H R, FABER A W, VOS J C, RAUÉ H A . Rrp5p, a trans-acting factor in yeast ribosome biogenesis, is an RNA-binding protein with a pronounced preference for U-rich sequences. RNA, 2006,12(2):263-271. |
| [8] | YAN P, LUO S, LU J Y, SHEN X . Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Current Opinion in Genetics and Development, 2017,46:170-178. |
| [9] | FAUQUENOY S, MIGEOT V, FINET O, YAGUE-SANZ C, KHOROSJUTINA O, EKWALL K, HERMAND D . Repression of cell differentiation by a cis-acting lincRNA in fission yeast. Current Biology, 2018,28(3):383-391. |
| [10] | SARVER A L, SUBRAMANIAN S . Competing endogenous RNA database. Bioinformation, 2012,8(15):731-733. |
| [11] | CESANA M, CACCHIARELLI D, LEGNINI I, SANTINI T, STHANDIER O, CHINAPPI M, TRAMONTANO A, BOZZONI I . A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell, 2011,147(2):358-369. |
| [12] | CHAN W L, HUANG H D, CHANG J G . LncRNAMap: A map of putative regulatory functions in the long non-coding transcriptome. Computational Biology and Chemistry, 2014,50:41-49. |
| [13] | KRISHNAN J, MISHRA R K . Emerging trends of long non-coding RNAs in gene activation. The FEBS Journal, 2014,281(1):34-45. |
| [14] | GUTTMAN M, AMIT I, GARBER M, FRENCH C, LIN M F, FELDSER D, HUARTE M, ZUK O, CAREY B W, CASSADY J P, CABILI M N, JAENISCH R, MIKKELSEN T S, JACKS T, HACOHEN N, BERNSTEIN B E, KELLIS M, REGEV A, RINN J L, LANDER E S . Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 2009,458(7235):223-227. |
| [15] | LIU J, JUNG C, XU J, WANG H, DENG S, BERNAD L, ARENAS-HUERTERO C, CHUA N H . Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. The Plant Cell, 2012,24(11):4333-4345. |
| [16] | CHEN B, ZHANG Y, ZHANG X, JIA S L, CHEN S, KANG L . Genome-wide identification and developmental expression profiling of long noncoding RNAs during Drosophila metamorphosis. Scientific Reports, 2016,6:23330. |
| [17] | LIU H, LI M Q, HE X Y, CAI S F, HE X K, LU X M . Transcriptome sequencing and characterization of ungerminated and germinated spores of Nosema bombycis. Acta Biochimica et Biophysica Sinica, 2016,48(3):246-256. |
| [18] | GUO R, CHEN D F, XIONG C L, HOU C S, ZHENG Y Z, FU Z M, DIAO Q Y, ZHANG L, WANG H Q, HOU Z X, LI W D, KUMAR D, LIANG Q . Identification of long non-coding RNAs in the chalkbrood disease pathogen Ascospheara apis. Journal of Invertebrate Pathology, 2018,156:1-5. |
| [19] | ATKINSON S R . The fission yeast non-coding transcriptome[D]. London: University College London, 2014. |
| [20] | GUO G J, KANG Q Z, CHEN Q H, JING O Y, CHEN J L . Abstract 3543: A long noncoding RNA plays a critical role in Bcr-Abl-mediated cellular transformation. Cancer Research, 2014,74(19):3543. |
| [21] | GUO R, CHEN D F, XIONG C L, HOU C S, ZHENG Y Z, FU Z M, LIANG Q, DIAO Q Y, ZHANG L, WANG H Q, HOU Z, KUMAR D . First identification of long non-coding RNAs in fungal parasite Nosema ceranae. Apidologie, 2018,49(5):660-670. |
| [22] | 郭睿, 耿四海, 熊翠玲, 郑燕珍, 付中民, 王海朋, 杜宇, 童新宇, 赵红霞, 陈大福 . 意大利蜜蜂工蜂中肠发育过程中长链非编码RNA的差异表达分析. 中国农业科学, 2018,51(18):3600-3613. |
| GUO R, GENG S H, XIONG C L, ZHENG Y Z, FU Z M, WANG H P, DU Y, TONG X Y, ZHAO H X, CHEN D F . Differential expression analysis of long non-coding RNAs during the developmental process of Apis mellifera ligustica worker’s midgut. Scientia Agricultura Sinica, 2018,51(18):3600-3613. (in Chinese) | |
| [23] | LI J, MA W, ZENG P, WANG J, GENG B, YANG J, CUI Q . LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Briefings in Bioinformatics, 2015,16(5):806-812. |
| [24] | SMOOT M E, ONO K, RUSCHEINSKI J, WANG P L, IDEKER T . Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics, 2011,27(3):431-432. |
| [25] | 郭睿, 杜宇, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 王海朋, 陈华枝, 耿四海, 周丁丁, 石彩云, 赵红霞, 陈大福 . 意大利蜜蜂幼虫肠道发育过程中的差异表达microRNA及其调控网络. 中国农业科学, 2018,51(21):4197-4209. |
| GUO R, DU Y, XIONG C L, ZHENG Y Z, FU Z M, XU G J, WANG H P, CHEN H Z, GENG S H, ZHOU D D, SHI C Y, ZHAO H X, CHEN D F . Differentially expressed microRNA and their regulation networks during the developmental process of Apis mellifera ligustica larval gut. Scientia Agricultura Sinica, 2018,51(21):4197-4209. (in Chinese) | |
| [26] | 杜宇, 童新宇, 周丁丁, 陈大福, 熊翠玲, 郑燕珍, 徐国钧, 王海朋, 陈华枝, 郭意龙, 隆琦, 郭睿 . 中华蜜蜂幼虫肠道响应球囊菌胁迫的microRNA应答分析. 微生物学报, 2019,59(9):1747-1764. |
| DU Y, TONG X Y, ZHOU D D, CHEN D F, XIONG C L, ZHENG Y Z, XU G J, WANG H P, CHEN H Z, GUO Y L, LONG Q, GUO R . MicroRNA responses in the larval gut of Apis cerana cerana to Ascosphaera apis stress. Acta Microbiologica Sinica, 2019,59(9):1747-1764. (in Chinese) | |
| [27] | BI F C, MENG X C, MA C, YI G J . Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genomics, 2015,16:776. |
| [28] | CASSELLI T, QURESHI H, PETERSON E, PERLEY D, BLAKE E, JOKINEN B, ABBAS A, NECHAEV S, WATT J A, DHASARATHY A, BRISSETTE C A . MicroRNA and mRNA transcriptome profiling in primary human astrocytes infected with Borrelia burgdorferi. PLoS ONE, 2017,12(1):e0170961. |
| [29] | JIANG X F, QIAO F, LONG Y L, CONG H Q, SUN H P . MicroRNA- like RNAs in plant pathogenic fungus Fusarium oxysporum f. sp. niveum are involved in toxin gene expression fine tuning. 3 Biotech, 2017, 7(5): Article 354. |
| [30] | FANG S S, ZHANG L L, GUO J C, NIU Y W, WU Y, LI H, ZHAO L H, LI X Y, TENG X Y, SUN X H, SUN L, ZHANG M Q, CHEN R S, ZHAO Y . NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Research, 2018,46(Database issue):D308-D314. |
| [31] | QUEK X C, THOMSON D W, MAAG J L V, BARTONICEK N, SIGNAL B, CLARK M B, GLOSS B S, DINGER M E . LncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Research, 2015,43(Database issue):D168-D173. |
| [32] | HARROW J, FRANKISH A, GONZALEZ J M, TAPANARI E, DIEKHANS M, KOKOCINSKI F, AKEN B L, BARRELL D, ZADISSA A, SEARLE S, BARNES I, BIGNELL A, BOYCHENKO V, HUNT T, KAY M, MUKHERJEE G, RAJAN J, DESPACIO- REYES G, SAUNDERS G, STEWARD C, HARTE R, LIN M, HOWALD C, TANZER A, DERRIEN T, CHRAST J, WALTERS N, BALASUBRAMANIAN S, PEI B, TRESS M, RODRIGUEZ J M, EZKURDIA I, VAN BAREN J, BRENT M, HAUSSLER D, KELLIS M, VALENCIA A, REYMOND A, GERSTEIN M, GUIGÓ R, HUBBARD T J . GENCODE: The reference human genome annotation for the ENCODE Project. Genome Research, 2012,22(9):1760-1774. |
| [33] | GUO R, CAO G L, LU Y H, XUE R Y, KUMAR D, HU X L, GONG C L . Exogenous gene can be integrated into Nosema bombycis genome by mediating with a non-transposon vector. Parasitology Research, 2016,115(8):3093-3098. |
| [34] | PALDI N, GLICK E, OLIVA M, ZILBERBERG Y, AUBIN L, PETTIS J, CHEN Y P, EVANS J D . Effective gene silencing in a microsporidian parasite associated with honeybee ( Apis mellifera) colony declines. Applied and Environmental Microbiology, 2010,76(17):5960-5964. |
| [35] |
HUANG Q, LI W, CHEN Y P, RETSCHNIG-TANNER G, YANEZ O, NEUMANN P, EVANS J D . Dicer regulates Nosema ceranae proliferation in honeybees. Insect Molecular Biology, 2019,28(1):74-85.
doi: 10.1111/imb.v28.1 |
| [36] |
WANG K C, CHANG H Y . Molecular mechanisms of long noncoding RNAs. Molecular Cell, 2011,43(6):904-914.
doi: 10.1016/j.molcel.2011.08.018 |
| [37] | ZHAN S Y, DONG Y, ZHAO W, GUO J Z, ZHONG T, WANG L J, LI L, ZHANG H P . Genome-wide identification and characterization of long non-coding RNAs in developmental skeletal muscle of fetal goat. BMC Genomics, 2016,17:666. |
| [38] | FRANZEN C, MULLER A, HARTMANN P, SALZBERGER B . Cell invasion and intracellular fate of Encephalitozoon cuniculi (Microsporidia). Parasitology, 2005,130(3):285-292. |
| [39] |
CHEN D F, CHEN H Z, DU Y, ZHOU D D, GENG S H, WANG H P, WAN J Q, XIONG C L, ZHENG Y Z, GUO R . Genome-wide identification of long non-coding RNAs and their regulatory networks involved in Apis mellifera ligustica response to Nosema ceranae infection. Insects, 2019,10(8):245.
doi: 10.3390/insects10080245 |
| [40] | YANG D L, PAN L X, CHEN Z Z, DU H H, LUO B, LUO J, PAN G Q . The roles of microsporidia spore wall proteins in the spore wall formation and polar tube anchorage to spore wall during development and infection processes. Experimental Parasitology, 2018,187:93-100. |
| [41] | GISDER S, MÖCKEL N, LINDE A, GENERSCH E . A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environmental Microbiology, 2011,13(2):404-413. |
| [42] | NAKANISHI N, OZAWA K, YAMADA S . Enzymes of the glycolytic pathway-phosphofructokinase, pyruvate kinase and lactate dehydrogenase// Dynamic Aspects of Dental Pulp, 1990: 203-220. |
| [43] |
DOLGIKH V V, SOKOLOVA J J, ISSI I V . Activities of enzymes of carbohydrate and energy metabolism of the spores of the microsporidian, Nosema grylli. The Journal of Eukaryotic Microbiology, 1997,44(3):246-249.
doi: 10.1111/jeu.1997.44.issue-3 |
| [44] |
ZIMMERMANN H . Extracellular ATP and other nucleotides— ubiquitous triggers of intercellular messenger release. Purinergic Signalling, 2015,12(1):25-57.
doi: 10.1007/s11302-015-9483-2 |
| [45] | HEINZ E, HACKER C, DEAN P, MIFSUD J, GOLDBERG A V, WILLIAMS T A, NAKJANG S, GREGORY A, HIRT R P, LUCOCQ J M, KUNJI E R S, EMBLEY T M . Plasma membrane-located purine nucleotide transport proteins are key components for host exploitation by microsporidian intracellular parasites. PLoS Pathogens, 2014,10(12):e1004547. |
| [46] |
SALMENA L, POLISENO L, TAY Y, KATS L, PANDOLFI P P . A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell, 2011,146(3):353-358.
doi: 10.1016/j.cell.2011.07.014 |
| [47] |
POLISENO L, SALMENA L, ZHANG J, CARVER B, HAVEMAN W J, PANDOLFI P P . A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 2010,465(7301):1033-1038.
doi: 10.1038/nature09144 |
| [48] |
WANG W T, YE H, WEI P P, HAN B W, HE B, CHEN Z H, CHEN Y Q . LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. Journal of Hematology and Oncology, 2016,9:117.
doi: 10.1186/s13045-016-0348-0 |
| [49] |
郭睿, 陈华枝, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 杜宇, 王海朋, 耿四海, 周丁丁, 刘思亚, 陈大福 . 意大利蜜蜂工蜂中肠发育过程中的差异表达环状RNA及其调控网络分析. 中国农业科学, 2018,51(23):4575-4590.
doi: 10.3864/j.issn.0578-1752.2018.23.015 |
|
GUO R, CHEN H Z, XIONG C L, ZHENG Y Z, FU Z M, XU G J, DU Y, WANG H P, GENG S H, ZHOU D D, LIU S Y, CHEN D F . Analysis of differentially expressed circular RNAs and their regulation networks during the developmental process of Apis mellifera ligustica worker’s midgut. Scientia Agricultura Sinica, 2018,51(23):4575-4590. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2018.23.015 |
| [1] | 冯睿蓉,付中民,杜宇,张文德,范小雪,王海朋,万洁琦,周紫彧,康育欣,陈大福,郭睿,史培颖. 中华蜜蜂幼虫肠道中微小RNA的鉴定及分析[J]. 中国农业科学, 2022, 55(1): 208-218. |
| [2] | 杜宇,范小雪,蒋海宾,王杰,冯睿蓉,张文德,余岢骏,隆琦,蔡宗兵,熊翠玲,郑燕珍,陈大福,付中民,徐国钧,郭睿. 微小RNA介导意大利蜜蜂工蜂对东方蜜蜂微孢子虫的跨界调控[J]. 中国农业科学, 2021, 54(8): 1805-1820. |
| [3] | 陈华枝,范元婵,蒋海宾,王杰,范小雪,祝智威,隆琦,蔡宗兵,郑燕珍,付中民,徐国钧,陈大福,郭睿. 基于纳米孔全长转录组数据完善东方蜜蜂微孢子虫的基因组注释[J]. 中国农业科学, 2021, 54(6): 1288-1300. |
| [4] | 陈华枝,王杰,祝智威,蒋海宾,范元婵,范小雪,万洁琦,卢家轩,郑燕珍,付中民,徐国钧,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中长链非编码RNA的比较及潜在功能分析[J]. 中国农业科学, 2021, 54(2): 435-448. |
| [5] | 王继卿,郝志云,沈继源,柯娜,黄兆春,梁维炜,罗玉柱,胡江,刘秀,李少斌. 小尾寒羊泌乳性状重要lncRNAs的筛选、鉴定及功能分析[J]. 中国农业科学, 2021, 54(14): 3113-3123. |
| [6] | 周丁丁, 范元婵, 王杰, 蒋海宾, 祝智威, 范小雪, 陈华枝, 杜宇, 周紫彧, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 蜜蜂球囊菌中长链非编码RNA的调控作用[J]. 中国农业科学, 2021, 54(1): 224-238. |
| [7] | 贾海燕,宋丽云,徐翔,解屹,张超群,刘天波,赵存孝,申莉莉,王杰,李莹,王凤龙,杨金广. 不同温度下TMV侵染枯斑三生烟的LncRNA差异表达[J]. 中国农业科学, 2020, 53(7): 1381-1396. |
| [8] | 耿四海,石彩云,范小雪,王杰,祝智威,蒋海宾,范元婵,陈华枝,杜宇,王心蕊,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 微小RNA介导东方蜜蜂微孢子虫侵染意大利蜜蜂工蜂的分子机制[J]. 中国农业科学, 2020, 53(15): 3187-3204. |
| [9] | 杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿. 微小RNA及其介导的竞争性内源RNA调控网络在意大利蜜蜂工蜂中肠发育过程中的潜在作用[J]. 中国农业科学, 2020, 53(12): 2512-2526. |
| [10] | 杜宇,周丁丁,万洁琦,卢家轩,范小雪,范元婵,陈恒,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 意大利蜜蜂工蜂中肠发育过程中的差异基因 表达谱及调控网络[J]. 中国农业科学, 2020, 53(1): 201-212. |
| [11] | 薛亚东,杨露,杨慧丽,李冰,林亚楠,张怀胜,郭战勇,汤继华. 玉米C型细胞质雄性不育花药不同发育时期的转录组分析[J]. 中国农业科学, 2019, 52(8): 1308-1323. |
| [12] | 付中民,陈华枝,刘思亚,祝智威,范小雪,范元婵,万洁琦,张璐,熊翠玲,徐国钧,陈大福,郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答[J]. 中国农业科学, 2019, 52(17): 3069-3082. |
| [13] | 郭睿,杜宇,童新宇,熊翠玲,郑燕珍,徐国钧,王海朋,耿四海,周丁丁,郭意龙,吴素珍,陈大福. 意大利蜜蜂幼虫肠道在球囊菌侵染前期的 差异表达microRNA及其调控网络[J]. 中国农业科学, 2019, 52(1): 166-180. |
| [14] | 郭睿,陈华枝,熊翠玲,郑燕珍,付中民,徐国钧,杜宇,王海朋,耿四海,周丁丁,刘思亚,陈大福. 意大利蜜蜂工蜂中肠发育过程中的差异表达环状RNA及其调控网络分析[J]. 中国农业科学, 2018, 51(23): 4575-4590. |
| [15] | 郭睿,杜宇,熊翠玲,郑燕珍,付中民,徐国钧,王海朋,陈华枝,耿四海,周丁丁,石彩云,赵红霞,陈大福. 意大利蜜蜂幼虫肠道发育过程中的差异表达 microRNA及其调控网络[J]. 中国农业科学, 2018, 51(21): 4197-4209. |
|
||