













中国农业科学 ›› 2022, Vol. 55 ›› Issue (10): 1938-1948.doi: 10.3864/j.issn.0578-1752.2022.10.005
李晓菁1( ),张思雨1,刘迪1,袁晓伟2,李兴盛2,石延霞1,谢学文1,李磊1,范腾飞1,李宝聚1(
),张思雨1,刘迪1,袁晓伟2,李兴盛2,石延霞1,谢学文1,李磊1,范腾飞1,李宝聚1( ),柴阿丽1(
),柴阿丽1( )
)
收稿日期:2021-11-18
接受日期:2021-12-20
出版日期:2022-05-16
发布日期:2022-06-02
通讯作者:
李宝聚,柴阿丽
作者简介:李晓菁,E-mail: 基金资助:
LI XiaoJing1( ),ZHANG SiYu1,LIU Di1,YUAN XiaoWei2,LI XingSheng2,SHI YanXia1,XIE XueWen1,LI Lei1,FAN TengFei1,LI BaoJu1(
),ZHANG SiYu1,LIU Di1,YUAN XiaoWei2,LI XingSheng2,SHI YanXia1,XIE XueWen1,LI Lei1,FAN TengFei1,LI BaoJu1( ),CHAI ALi1(
),CHAI ALi1( )
)
Received:2021-11-18
Accepted:2021-12-20
Online:2022-05-16
Published:2022-06-02
Contact:
BaoJu LI,ALi CHAI
摘要:
【目的】由芸薹根肿菌(Plasmodiophora brassicae)侵染引起的十字花科根肿病是一种世界性土传病害,病原菌长期存在于土壤中,对十字花科作物造成严重威胁。改良叠氮溴化丙锭(propidium monoazide xx,PMAxx)可选择性地穿透受损的死细胞膜,并抑制死细胞DNA的实时荧光定量PCR(qPCR)扩增。本文将PMAxx与qPCR技术相结合,建立一种快速检测芸薹根肿菌活菌的方法,为根肿病的早期诊断及制定科学的防控措施提供依据。【方法】配置浓度分别为0、5、10、20、40、60 µmol·L-1的叠氮溴化丙锭PMA和改良叠氮溴化丙锭PMAxx,比较两种核酸染料对芸薹根肿菌死细胞DNA扩增的抑制效果,确定最佳核酸染料及工作浓度;设置光照时间分别为0、2、5、10、15和20 min,进行最佳光照时间的优化,建立芸薹根肿菌活细胞PMAxx-qPCR快速检测体系。设置芸薹根肿菌活孢子百分比为0、0.01%、0.1%、1%、10%、25%、50%、75%和100%的混合体系,验证PMAxx-qPCR体系的准确性,并应用于田间土壤样本中芸薹根肿菌活孢子的定量检测。【结果】PMAxx对芸薹根肿菌死细胞DNA的扩增抑制效果更好,当芸薹根肿菌浓度为1×108个孢子/mL,PMAxx预处理的最适终浓度为4 µmol·L-1,最佳光照时间为10 min时,可有效地抑制死孢子DNA的扩增,仅以有活力孢子DNA为靶标选择性地扩增。利用PMAxx-qPCR技术检测已知不同活孢子比例的菌悬液样品,各样品实测孢子存活率和理论存活率之间呈正相关(R2=0.992)。对田间采集的25份土壤样本,采用PMAxx-qPCR方法检测到11份样本中携带芸薹根肿菌,活细胞DNA浓度为32.35—6.97×103 fg·g-1。【结论】建立了基于PMAxx-qPCR的芸薹根肿菌活细胞定量检测技术,该技术具有快速、准确、灵敏的特点,解决了qPCR不能仅对活体病原菌进行准确鉴别和定量分析的问题,为制定有效的根肿病防控策略提供了依据。
李晓菁,张思雨,刘迪,袁晓伟,李兴盛,石延霞,谢学文,李磊,范腾飞,李宝聚,柴阿丽. 芸薹根肿菌活细胞PMAxx-qPCR快速定量检测方法的建立与应用[J]. 中国农业科学, 2022, 55(10): 1938-1948.
LI XiaoJing,ZHANG SiYu,LIU Di,YUAN XiaoWei,LI XingSheng,SHI YanXia,XIE XueWen,LI Lei,FAN TengFei,LI BaoJu,CHAI ALi. Establishment and Application of Rapid Quantitative Detection of Viable Plasmodiophora brassicae by PMAxx-qPCR Method[J]. Scientia Agricultura Sinica, 2022, 55(10): 1938-1948.
表1
引物特异性检测所用菌株"
| 序号No. | 病原菌Pathogen | 菌株编号Strain code | 寄主Host | 采集地Geographic origin |
|---|---|---|---|---|
| 1 | 芸薹根肿菌Plasmodiophora brassicae | BC18052103 | 大白菜Chinese cabbage | 云南省昆明市Kunming, Yunnan |
| 2 | 尖镰孢Fusarium oxysporum | GL17061203 | 甘蓝 Cabbage | 陕西省太白县Taibai, Shaanxi |
| 3 | 茄镰孢Fusarium solani | BC10120305 | 大白菜Chinese cabbage | 辽宁省丹东市Dandong, Liaoning |
| 4 | 半裸镰孢Fusarium incarnatum | HYC15171302 | 花椰菜Broccoli | 河北省承德市Chengde, Hebei |
| 5 | 立枯丝核菌Rhizoctonia solani | BC16072809 | 普通白菜Pakchoi | 河北省张家口市Zhangjiakou, Hebei |
| 6 | 核盘菌Sclerotinia sclerotiorum | YC19081601 | 油菜Rape | 山东省临沂市Linyi, Shandong |
| 7 | 芸薹链格孢Alternaria brassicae | BC15110628 | 大白菜Chinese cabbage | 河北省廊坊市Langfang, Hebei |
| 8 | 长孢轮枝菌Verticillium longisporum | BC13091524 | 大白菜Chinese cabbage | 河北省张家口市Zhangjiakou, Hebei |
| 9 | 瓜果腐霉Pythium aphanidermatum | BC12073101 | 大白菜Chinese cabbage | 江苏常州市Changzhou, Jiangsu |
| 10 | 灰葡萄孢Botrytis cinerea | HYC20051608 | 花椰菜Broccoli | 北京市顺义区Shunyi, Beijing |
| 11 | 胡萝卜软腐果胶杆菌胡萝卜亚种 Pectobacterium carotovorun subsp. carotovorum | BC14120625 | 大白菜Chinese cabbage | 内蒙古自治区乌兰察布市 Ulanqab, Inner Mongolia |
| 12 | 野油菜黄单胞菌野油菜致病变种 Xanthomonas campestris pv. campestris | GL18080621 | 甘蓝 Cabbage | 湖南省长沙市Changsha, Hunan |
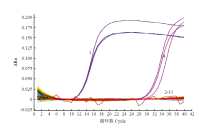
图1
荧光定量PCR引物特异性验证扩增曲线 1:芸薹根肿菌P. brassicae;2:尖镰孢F. oxysporum;3:茄镰孢F. solani;4:半裸镰孢F. incarnatum;5:立枯丝核菌R. solani;6:核盘菌S. sclerotiorum;7:芸薹链格孢A. brassicae;8:长孢轮枝菌V. longisporum;9:瓜果腐霉P. aphanidermatum;10:灰葡萄孢B. cinerea;11:胡萝卜软腐果胶杆菌胡萝卜亚种P. carotovorun subsp. carotovorum;12:野油菜黄单胞菌野油菜致病变种X. campestris pv. campestris;13:健康大白菜根组织Root tissue of healthy Chinese cabbage;14:ddH2O"
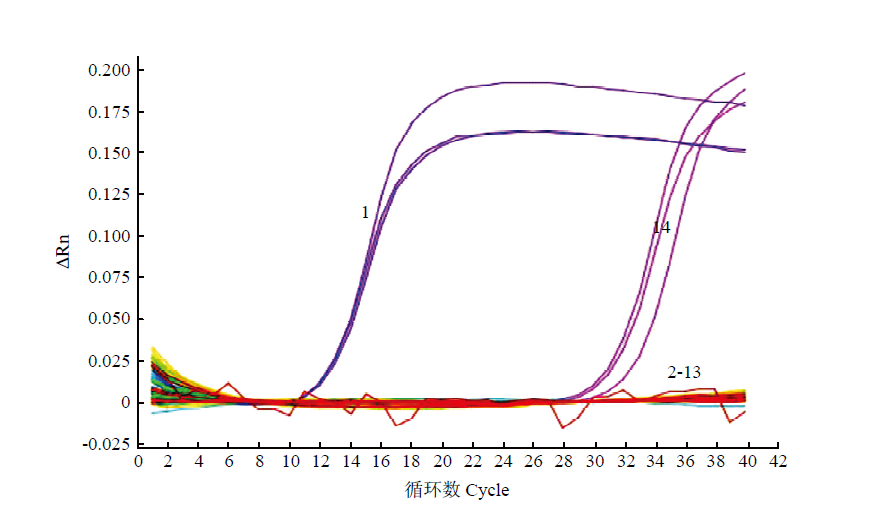
表2
PMA和PMAxx预处理芸薹根肿菌qPCR检测的Ct值"
| 光敏染料 Photoactivatable dye | 浓度 Concentration (µmol∙L-1) | Ct (活孢子Viable spores) | Ct (死孢子Dead spores) | dCt (活孢子Viable spores) | dCt (死孢子Dead spores) | ddCt |
|---|---|---|---|---|---|---|
| PMA/PMAxx | 0 | 14.53a | 14.86a | / | / | / |
| PMA | 5 | 15.53b | 25.82b | 1.00 | 10.96 | 9.96 |
| 10 | 15.90b | 27.43c | 1.37 | 12.57 | 11.20 | |
| 20 | 16.84c | 28.92d | 2.31 | 14.06 | 11.75 | |
| 40 | 17.90d | 29.71e | 3.37 | 14.85 | 11.48 | |
| 60 | 18.52e | 29.84e | 3.99 | 14.98 | 10.99 | |
| PMAxx | 5 | 15.56b | 27.91c | 1.03 | 13.05 | 12.02 |
| 10 | 16.21bc | 29.57d | 1.68 | 14.71 | 13.03 | |
| 20 | 17.18c | 30.59e | 2.65 | 15.73 | 13.08 | |
| 40 | 18.21d | 32.10f | 3.68 | 17.24 | 13.56 | |
| 60 | 18.58e | 32.61f | 4.05 | 17.75 | 13.70 |
表3
不同比例芸薹根肿菌活孢子悬浮液qPCR和PMAxx-qPCR检测结果"
| 活孢子百分比 Ratio of viable spores (%) | 活孢子浓度 Concentration of viable spores (spores/mL) | qPCR | PMAxx-qPCR | ||
|---|---|---|---|---|---|
| Ct值 Ct value | DNA浓度对数值 Lg DNA concentration | Ct值 Ct value | DNA浓度对数值 Lg DNA concentration | ||
| 100 | 1.00×108 | 10.04±0.28 | 6.74±0.08a | 10.34±0.21 | 6.66±0.06a |
| 75 | 7.50×107 | 10.76±0.56 | 6.58±0a | 11.10±0.24 | 6.45±0.07b |
| 50 | 5.00×107 | 10.26±0.23 | 6.72±0.12a | 12.13±0.47 | 6.16±0.13c |
| 25 | 2.50×107 | 10.48±0.65 | 6.66±0.10a | 13.31±0.37 | 5.83±0.10d |
| 10 | 1.00×107 | 10.19±0.53 | 6.74±0.10a | 14.66±0.25 | 5.46±0.07e |
| 1 | 1.00×106 | 10.40±0.39 | 6.68±0.14a | 18.01±0.39 | 4.53±0.11f |
| 0.1 | 1.00×105 | 10.81±0.62 | 6.57±0.13a | 20.63±0.49 | 3.80±0.13g |
| 0.01 | 1.00×104 | 10.57±0.52 | 6.63±0.10a | 23.59±0.34 | 2.98±0.09h |
表4
十字花科根肿病田间发病土壤带菌量qPCR及PMAxx-qPCR检测结果"
| 样本 Sample No. | 采集地 Geographic origin | 种植作物 Crop | 采样时间Sampling time | qPCR | PMAxx-qPCR | 病情指数Disease index | ||
|---|---|---|---|---|---|---|---|---|
| Ct值 Ct value | 浓度<BOLD>C</BOLD>oncentration (fg DNA·g-1 soil) | Ct值 Ct value | 浓度<BOLD>C</BOLD>oncentration (fg DNA·g-1 soil) | |||||
| 1 | 四川省绵阳市 Mianyang, Sichuan | 甘蓝Cabbage | 2020-07 | 18.59±0.16 | 2.32×104 | 20.86±0.29 | 5.43×103 | 47.61±0.58 |
| 2 | 四川省绵阳市 Mianyang, Sichuan | 甘蓝Cabbage | 2020-07 | 20.65±0.26 | 6.23×103 | 22.62±0.34 | 1.76×103 | 38.20±0.30 |
| 3 | 四川省广元市 Guangyuan, Sichuan | 甘蓝Cabbage | 2020-08 | 21.12±1.36 | 4.61×103 | 23.50±0.25 | 1.01×103 | 32.10±0.44 |
| 4 | 四川省广元市 Guangyuan, Sichuan | 甘蓝Cabbage | 2020-08 | 23.59±0.31 | 9.52×102 | 25.48±0.28 | 2.84×102 | 26.11±0.52 |
| 5 | 湖北省恩施州 Enshi, Hubei | 大白菜Chinese cabbage | 2020-07 | 17.00±0.50 | 6.44×104 | 20.47±0.70 | 6.97×103 | 50.48±0.63 |
| 6 | 湖北省恩施州 Enshi, Hubei | 大白菜Chinese cabbage | 2020-07 | 19.95±0.89 | 9.74×103 | 20.95±0.53 | 5.14×103 | 44.85±0.63 |
| 7 | 山东省青岛市 Qingdao, Shandong | 大白菜Chinese cabbage | 2020-07 | 23.08±0.42 | 1.31×103 | 25.03±0.33 | 3.78×102 | 26.36±0.72 |
| 8 | 山东省青岛市 Qingdao, Shandong | 小白菜Pakchoi | 2020-07 | 25.21±0.89 | 3.37×102 | 28.88±0.30 | 32.35 | 0 |
| 9 | 河南省南阳市 Nanyang, Henan | 甘蓝Cabbage | 2020-08 | 20.68±0.77 | 6.12×103 | 24.55±0.41 | 7.86×102 | 27.94±0.58 |
| 10 | 江苏省无锡市 Wuxi, Jiangsu | 甘蓝Cabbage | 2020-09 | 24.80±0.67 | 4.39×102 | 26.71±0.23 | 1.29×102 | 20.07±0.32 |
| 11 | 辽宁省沈阳市 Shenyang, Liaoning | 大白菜Chinese cabbage | 2020-10 | 24.14±0.79 | 6.70×102 | 25.95±0.55 | 2.10×102 | 21.62±0.60 |
| 其余14份土壤样本 The other 14 samples | >35.00 | 0 | >35.00 | 0 | 0 | |||
| [1] |
DIXON G R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. Journal of Plant Growth Regulation, 2009, 28(3): 194-202.
doi: 10.1007/s00344-009-9090-y |
| [2] |
HOWARD R J, STRELKOV S E, HARDING M W. Clubroot of cruciferous crops - new perspectives on an old disease. Canadian Journal of Plant Pathology, 2010, 32(1): 43-57.
doi: 10.1080/07060661003621761 |
| [3] |
DEVOS S, VISSENBERG K, VERBELEN J P, PRINSEN E. Infection of Chinese cabbage by Plasmodiophora brassicae leads to a stimulation of plant growth: Impacts on cell wall metabolism and hormone balance. New Phytologist, 2005, 166(1): 241-250.
doi: 10.1111/j.1469-8137.2004.01304.x |
| [4] |
HWANG S F, STRELKOV S E, FENG J, GOSSEN B D, HOWARD R J. Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Molecular Plant Pathology, 2012, 13(2): 105-113.
doi: 10.1111/j.1364-3703.2011.00729.x |
| [5] |
TSO H H, GALINDO-GONZALEZ L, STRELKOV S E. Current and future pathotyping platforms for Plasmodiophora brassicae in Canada. Plants, 2021, 10(7): 1446.
doi: 10.3390/plants10071446 |
| [6] |
CHAI A L, XIE X W, SHI Y X, LI B J. Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Canadian Journal of Plant Pathology, 2014, 36: 142-153.
doi: 10.1080/07060661.2013.868829 |
| [7] |
DONALD C, PORTER I. Integrated control of clubroot. Journal of Plant Growth Regulation, 2009, 28: 289-303.
doi: 10.1007/s00344-009-9094-7 |
| [8] |
TSUSHIMA S. Perspective of integrated pest management - A case study: Clubroot disease of crucifers. Journal of Pesticide Science, 2000, 25(3): 296-299.
doi: 10.1584/jpestics.25.296 |
| [9] |
ITO S, MAEHARA T, MARUNO E, TANAKA S, KAMEYA-IWAKI M, KISHI F. Development of a PCR-based assay for the detection of Plasmodiophora brassicae in soil. Journal of Phytopathology, 1999, 147(2): 83-88.
doi: 10.1111/j.1439-0434.1999.tb03812.x |
| [10] | 杨佩文, 杨勤忠, 王群, 李家瑞, 曾莉. 十字花科蔬菜根肿病菌的PCR检测. 云南农业大学学报, 2002, 17(2): 137-139, 157. |
| YANG P W, YANG Q Z, WANG Q, LI J R, ZENG L. PCR detection of Plasmodiophora brassicae causing cruciferae clubroot. Journal of Yunnan Agricultural University, 2002, 17(2): 137-139, 157. (in Chinese) | |
| [11] |
CAO T, TEWARI J, STRELKOV S E. Molecular detection of Plasmodiophora brassicae, causal agent of clubroot of crucifers, in plant and soil. Plant Disease, 2007, 91(1): 80-87.
doi: 10.1094/PD-91-0080 |
| [12] |
FAGGIAN R, STRELKOV S E. Detection and measurement of Plasmodiophora brassicae. Journal of Plant Growth Regulation, 2009, 28: 282-288.
doi: 10.1007/s00344-009-9092-9 |
| [13] |
FAGGIAN R, BULMAN S R, LAWRIE A C, PORTER I J. Specific polymerase chain reaction primers for the detection of Plasmodiophora brassicae in soil and water. Phytopathology, 1999, 89(5): 392-397.
doi: 10.1094/PHYTO.1999.89.5.392 |
| [14] |
WALLENHAMMAR A C, ARWIDSSON O. Detection of Plasmodiophora brassicae by PCR in naturally infested soils. European Journal of Plant Pathology, 2001, 107(3): 313-321.
doi: 10.1023/A:1011224503200 |
| [15] | 李淼, 周丽洪, 刘雅婷, 刘峰, 杨俊, 姬广海. 云南省十字花科蔬菜根肿病的实时荧光定量PCR检测. 云南农业大学学报 (自然科学), 2016, 31(1): 43-48. |
| LI M, ZHOU L H, LIU Y T, LIU F, YANG J, JI G H. Detection of Plasmodiophora brassicae with real-time quantitative PCR in Yunnan Province. Journal of Yunnan Agricultural University (Natural Science), 2016, 31(1): 43-48. (in Chinese) | |
| [16] |
WALLENHAMMAR A C, ALMQUIST C, SÖDERSTRÖM M, JONSSON A. In-field distribution of Plasmodiophora brassicae measured using quantitative real-time PCR. Plant Pathology, 2012, 61(1): 16-28.
doi: 10.1111/j.1365-3059.2011.02477.x |
| [17] |
LI J P, LI Y, SHI Y X, XIE X W, CHAI A L, LI B J. Development of a real-time PCR assay for Plasmodiophora brassicae and its detection in soil samples. Journal of Integrative Agriculture, 2013, 12(10): 1799-1806.
doi: 10.1016/S2095-3119(13)60491-8 |
| [18] |
CHAI A L, LI J P, XIE X W, SHI Y X, LI B J. Dissemination of Plasmodiophora brassicae in livestock manure detected by qPCR. Plant Pathology, 2016, 65(1): 137-144.
doi: 10.1111/ppa.12391 |
| [19] |
WEN R, LEE J, CHU M, TONU N, DUMONCEAUX T, GOSSEN B D, YU F, PENG G. Quantification of Plasmodiophora brassicae resting spores in soils using droplet digital PCR (ddPCR). Plant Disease, 2020, 104(4): 1188-1194.
doi: 10.1094/PDIS-03-19-0584-RE |
| [20] |
RENNIE D C, MANOLII V P, CAO T, HWANG S F, HOWARD R J, STRELKOV S E. Direct evidence of surface infestation of seeds and tubers by Plasmodiophora brassicae and quantification of spore loads. Plant Pathology, 2011, 60(5): 811-819.
doi: 10.1111/j.1365-3059.2011.02449.x |
| [21] |
AL-DAOUD F, GOSSEN B D, ROBSON J, MCDONALD M R. Propidium monoazide improves quantification of resting spores of Plasmodiophora brassicae with qPCR. Plant Disease, 2017, 101(3): 442-447.
doi: 10.1094/PDIS-05-16-0715-RE |
| [22] |
NOCKER A, CAMPER A K. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Applied and Environmental Microbiology, 2006, 72(3): 1997-2004.
doi: 10.1128/AEM.72.3.1997-2004.2006 |
| [23] |
LIU Y, MUSTAPHA A. Detection of viable Escherichia coli O157: H7 in ground beef by propidium monoazide real-time PCR. International Journal of Food Microbiology, 2014, 170: 48-54.
doi: 10.1016/j.ijfoodmicro.2013.10.026 |
| [24] |
WANG S, LEVIN R E. Discrimination of viable Vibrio vulnificus cells from dead cells in real-time PCR. Journal of Microbiological Methods, 2006, 64(1): 1-8.
doi: 10.1016/j.mimet.2005.04.023 |
| [25] |
YÁÑEZ M A, NOCKER A, SORIA-SORIA E, MÚRTULA R, MARTÍNEZ L, CATALÁN V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. Journal of Microbiological Methods, 2011, 85(2): 124-130.
doi: 10.1016/j.mimet.2011.02.004 |
| [26] |
CRESPO-SEMPERE A, ESTIARTE N, MARÍN S, SANCHIS V, RAMOS A J. Propidium monoazide combined with real-time quantitative PCR to quantify viable Alternaria spp. contamination in tomato products. International Journal of Food Microbiology, 2013, 165(3): 214-220.
doi: 10.1016/j.ijfoodmicro.2013.05.017 |
| [27] |
HONG W, XIONG J, NYARUABA R, LI J H, MUTURI E, LIU H, YU J P, YANG H, WEI H P. Rapid determination of infectious SARS-CoV-2 in PCR-positive samples by SDS-PMA assisted RT-qPCR. Science of the Total Environment, 2021, 797: 149085.
doi: 10.1016/j.scitotenv.2021.149085 |
| [28] |
LUO L X, WALTERS C, BOLKAN H, LIU X L, LI J Q. Quantification of viable cells of Clavibacter michiganensis subsp. michiganensis using a DNA binding dye and a real-time PCR assay. Plant Pathology, 2008, 57(2): 332-337.
doi: 10.1111/j.1365-3059.2007.01736.x |
| [29] |
MENG X L, CHAI A L, CHEN L, SHI Y X, XIE X W, MA Z H, LI B J. Rapid detection and quantification of viable Pseudomonas syringae pv. lachrymans cells in contaminated cucumber seeds using propidium monoazide and a real-time PCR assay. Canadian Journal of Plant Pathology, 2016, 38(3): 296-306.
doi: 10.1080/07060661.2016.1216897 |
| [30] |
TIAN Q, FENG J J, HU J, ZHAO W J. Selective detection of viable seed-borne Acidovorax citrulli by real-time PCR with propidium monoazide. Scientific Reports, 2016, 6: 35457.
doi: 10.1038/srep35457 |
| [31] |
HAN S N, JIANG N, LV Q Y, KAN Y M, HAO J J, LI J Q, LUO L X. Detection of Clavibacter michiganensis subsp. michiganensis in viable but nonculturable state from tomato seed using improved qPCR. PLoS ONE, 2018, 13(5): e0196525.
doi: 10.1371/journal.pone.0196525 |
| [32] |
CHAI A L, BEN H Y, GUO W T, SHI Y X, XIE X W, LI L, LI B J. Quantification of viable cells of Pseudomonas syringae pv. tomato in tomato seed using propidium monoazide and a real-time PCR assay. Plant Disease, 2020, 104(8): 2225-2232.
doi: 10.1094/PDIS-11-19-2397-RE pmid: 32452750 |
| [1] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [2] | 朱春艳,宋佳伟,白天亮,王娜,马帅国,普正菲,董艳,吕建东,李杰,田蓉蓉,罗成科,张银霞,马天利,李培富,田蕾. NaCl胁迫对不同耐盐性粳稻种质幼苗叶绿素荧光特性的影响[J]. 中国农业科学, 2022, 55(13): 2509-2525. |
| [3] | 许晨,王文静,曹珊,李如雪,张贝贝,孙爱清,张春庆. 花后DA-6处理调控小麦种子活力的机理[J]. 中国农业科学, 2021, 54(9): 1821-1834. |
| [4] | 李天聪,朱行,魏宁,龙凤,武建颖,张燕,董金皋,申珅,郝志敏. 玉米大斑病菌SC35同源基因表达规律与互作分析[J]. 中国农业科学, 2021, 54(4): 733-743. |
| [5] | 赵立群,邱艳红,张晓飞,刘慧,杨静静,张建,张海军,徐秀兰,温常龙. TaqMan探针法实时荧光定量PCR检测西瓜潜隐病毒[J]. 中国农业科学, 2021, 54(20): 4337-4347. |
| [6] | 龙凤,王擎,朱行,王建霞,申珅,刘宁,郝志敏,董金皋. 玉米大斑病菌Septin基因家族的鉴定与表达模式分析[J]. 中国农业科学, 2020, 53(24): 5017-5026. |
| [7] | 康俊梅,张俏燕,蒋旭,王珍,张铁军,龙瑞才,崔会婷,杨青川. 紫花苜蓿MsSQE1的克隆及对皂甙合成的功能分析[J]. 中国农业科学, 2020, 53(2): 247-260. |
| [8] | 田媛,王力,龙凤,昝林森,成功. 人溶菌酶密码子优化及其在牛乳腺细胞中高效表达[J]. 中国农业科学, 2020, 53(18): 3805-3817. |
| [9] | 赵绪生,齐永志,甄文超. 小麦、玉米两熟区小麦纹枯病菌群体组成及其农田分布特征[J]. 中国农业科学, 2020, 53(16): 3269-3279. |
| [10] | 张道伟,康奎,余亚娅,匡富萍,潘碧莹,陈静,唐斌. 白背飞虱酚氧化酶原PPO基因特性及其免疫应答[J]. 中国农业科学, 2020, 53(15): 3108-3119. |
| [11] | 刘亦然,张虹,靳继苏,周忠实,郭建英. 莲草直胸跳甲Halloween基因家族鉴定及表达分析[J]. 中国农业科学, 2020, 53(10): 2009-2019. |
| [12] | 李文学, 肖瑞刚, 吕苗苗, 丁宁, 石华荣, 顾沛雯. 葡萄霜霉病菌实时荧光定量PCR检测体系的建立和应用[J]. 中国农业科学, 2019, 52(9): 1529-1540. |
| [13] | 丁艳娟,刘永康,罗雨嘉,邓颖梅,徐红星,唐斌,徐彩娣. 褐飞虱GSK-3调控糖原与海藻糖代谢的潜在功能[J]. 中国农业科学, 2019, 52(7): 1237-1246. |
| [14] | 唐斌,沈祺达,曾伯平,肖仲久,邱玲玉,潘碧莹,李昆,张道伟. 褐飞虱一个新的海藻糖合成酶基因的特性、 发育表达及RNAi效果分析[J]. 中国农业科学, 2019, 52(3): 466-477. |
| [15] | 彭军波,李兴红,张玮,周莹,黄金宝,燕继晔. 葡萄溃疡病菌外泌蛋白LtGH61A的致病力及基因表达模式[J]. 中国农业科学, 2019, 52(24): 4518-4526. |
|
||