













中国农业科学 ›› 2020, Vol. 53 ›› Issue (12): 2512-2526.doi: 10.3864/j.issn.0578-1752.2020.12.017
• 畜牧·兽医·资源昆虫 • 上一篇
杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿
收稿日期:2019-10-27
出版日期:2020-06-16
发布日期:2020-06-25
作者简介:杜宇,E-mail:m18505700830@163.com。|范小雪,E-mail:imfanxx@163.com。
基金资助:DU Yu,FAN XiaoXue,JIANG HaiBin,WANG Jie,FAN YuanChan,ZHU ZhiWei,ZHOU DingDing,WAN JieQi,LU JiaXuan,XIONG CuiLing,ZHENG YanZhen,CHEN DaFu,GUO Rui
Received:2019-10-27
Online:2020-06-16
Published:2020-06-25
摘要:
【目的】微小RNA(microRNA,miRNA)能够通过靶向结合导致mRNA的抑制或降解,从而对基因表达进行负调控。MiRNA在昆虫的生长、发育、免疫和细胞生命活动调控方面扮演关键角色。本研究旨在对意大利蜜蜂(Apis mellifera ligustica,简称意蜂)工蜂中肠的差异表达miRNA(differentially expressed miRNA,DEmiRNA)及其调控网络进行深入分析,并结合前期在mRNA、长链非编码RNA(long non-coding RNA,lncRNA)和环状RNA(circular RNA,circRNA)组学层面的相关研究结果,系统解析中肠发育过程的miRNA差异表达谱及竞争性内源RNA(competing endogenous RNA,ceRNA)调控网络介导的中肠发育机理。【方法】利用small RNA-seq技术对正常的意蜂7日龄和10日龄工蜂中肠(Am7和Am10)进行测序。将质控后的测序数据比对西方蜜蜂基因组,再将比对上的序列标签(tags)比对到miRBase数据库以鉴定已知miRNA。利用每百万标签序列(tags per million,TPM)算法对miRNA表达量进行计算和归一化处理。根据|log2fold change|≥1且P≤0.05的标准筛选得到显著性DEmiRNA;利用相关软件进行靶mRNA的预测及GO和KEGG数据库注释。根据靶向结合关系及前期研究结果,利用Cytoscape软件对AMPK、PI3K-Akt、Wnt、cAMP、Hippo、mTOR、Toll/Imd、TGF-beta和MAPK 9条信号通路相关的DElncRNA/DEcircRNA-DEmiRNA-DEmRNA网络进行可视化;构建以miR-342-y为核心的ceRNA调控网络,进一步对网络中的靶mRNA进行代谢通路注释。利用茎环实时荧光定量PCR(Stem-loop RT-qPCR)验证测序数据及miRNA差异表达的可靠性。【结果】Am7 vs Am10比较组中共有112个显著性DEmiRNA,包括38个显著上调miRNA和74个显著下调miRNA,分别靶向结合7 434和9 559个mRNA,它们可注释到生物学进程相关的21和23个功能条目,细胞组分相关的16和17个功能条目,以及分子功能相关的10和11个功能条目;此外还能注释到果糖和甘露糖代谢、嘌呤代谢、甘氨酸、丝氨酸和苏氨酸代谢等物质代谢相关的83和86条通路;内吞作用、泛素蛋白水解、黑色素生成等细胞免疫相关的10和10条通路;MAPK、Jak-STAT、NF-κB等体液免疫相关的5和5条通路;以及Hippo、FoxO、Notch等涉及生长发育的13和11条信号通路。进一步构建上述9条信号通路相关的ceRNA调控网络,分析发现意蜂中肠发育过程中DEmiRNA与DElncRNA、DEcircRNA和DEmRNA之间存在复杂的调控关系;DEmiRNA位于网络中心,而DElncRNA、DEcircRNA和DEmRNA位于网络的外周。miR-342-y在意蜂工蜂中肠和幼虫肠道发育过程中均为显著性下调表达,可靶向结合3个DEcircRNA、4个DElncRNA和327个mRNA。Stem-loop RT-qPCR结果显示4个DEmiRNA的差异表达趋势与测序结果一致,说明本研究中测序数据及miRNA差异表达趋势真实可靠。【结论】DEmiRNA可能通过参与调控物质和能量代谢通路、Hippo和Wnt等信号通路以及细胞和体液免疫通路相关基因的表达影响意蜂中肠的生长和发育;miR-182-x、miR-291-y、miR-342-y、ame-miR-6001-3p等关键DEmiRNA介导的ceRNA调控网络可能在意蜂中肠发育过程发挥重要的调控作用。
杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿. 微小RNA及其介导的竞争性内源RNA调控网络在意大利蜜蜂工蜂中肠发育过程中的潜在作用[J]. 中国农业科学, 2020, 53(12): 2512-2526.
DU Yu,FAN XiaoXue,JIANG HaiBin,WANG Jie,FAN YuanChan,ZHU ZhiWei,ZHOU DingDing,WAN JieQi,LU JiaXuan,XIONG CuiLing,ZHENG YanZhen,CHEN DaFu,GUO Rui. The Potential Role of MicroRNAs and MicroRNA-Mediated Competing Endogenous Networks During the Developmental Process of Apis mellifera ligustica Worker’s Midgut[J]. Scientia Agricultura Sinica, 2020, 53(12): 2512-2526.
表1
RT-qPCR引物信息"
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) |
|---|---|
| Loop-miR-210-z | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGCCGC |
| F-miR-210-z | ACACTCCAGCTGGGCTGTGCGTGTGACA |
| Loop-miR-342-y | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACGGGTGC |
| F-miR-342-y | ACACTCCAGCTGGGTCTCACACAGAAATC |
| Loop-miR-7975-y | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGTGCCG |
| F-miR-7975-y | ACACTCCAGCTGGGATCCTGGTCA |
| Loop-miR-155-x | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACCCCTA |
| F-miR-155-x | ACACTCCAGCTGGGTTAATGCTAATTGTGA |
| R | CTCAACTGGTGTCGTGGA |
| U6-F | GTTAGGCTTTGACGATTTCG |
| U6-R | GGCATTTCTCCACCAGGTA |
表3
意蜂工蜂中肠发育过程中显著上调前10个miRNA"
| 差异表达miRNA ID DEmiRNA ID | 以2为底miRNA的相对变化倍数的对数值 log2fold change | P值 P value |
|---|---|---|
| miR-7132-y | 13.27 | 8.68E-09 |
| miR-2188-x | 12.72 | 2.31E-06 |
| miR-1388-x | 12.43 | 6.78E-06 |
| miR-1338-x | 12.38 | 7.39E-06 |
| miR-1388-y | 11.76 | 4.20E-04 |
| miR-1338-y | 11.67 | 9.18E-04 |
| miR-7132-x | 11.16 | 9.01E-03 |
| miR-3964-y | 11.09 | 3.37E-03 |
| miR-7935-y | 11.01 | 4.31E-03 |
| miR-6537-x | 10.46 | 0.023 |
表4
意蜂工蜂中肠发育过程中显著下调前10个miRNA"
| 差异表达miRNA ID DEmiRNA ID | 以2为底miRNA的相对变化倍数的对数值 log2fold change | P值 P value |
|---|---|---|
| miR-8503-x | -14.09 | 3.09E-04 |
| miR-298-x | -13.64 | 2.65E-08 |
| miR-291-y | -12.94 | 8.67E-05 |
| miR-292-y | -12.75 | 3.40E-04 |
| miR-4700-x | -12.55 | 0.015 |
| miR-293-y | -12.27 | 6.77E-04 |
| miR-2965-y | -12.13 | 0.030 |
| miR-2518-y | -12.13 | 6.13E-03 |
| miR-300-y | -12.03 | 2.96E-04 |
| miR-4577-y | -12.01 | 0.038 |
表5
意蜂工蜂中肠上调miRNA的靶mRNA显著富集的前15位通路"
| 通路 Pathway | 通路ID Pathway ID | 靶mRNA富集数 Number of target mRNAs | P值 P value |
|---|---|---|---|
| 钙信号通道 Calcium signaling pathway | ko04020 | 77 | 1.30E-14 |
| 催产素信号通路 Oxytocin signaling pathway | ko04921 | 82 | 1.07E-14 |
| 坏死性凋亡 Necroptosis | ko04217 | 58 | 5.72E-16 |
| 嗅觉传导 Olfactory transduction | ko04740 | 50 | 3.01E-17 |
| 长时程增强 Long-term potentiation | ko04720 | 70 | 3.98E-18 |
| Hippo信号通路 Hippo signaling pathway | ko04390 | 118 | 1.29E-18 |
| 促性腺激素释放激素信号通路 GnRH signaling pathway | ko04912 | 79 | 4.76E-19 |
| 神经营养因子信号通路 Neurotrophin signaling pathway | ko04722 | 89 | 3.17E-21 |
| 胆碱能突触 Cholinergic synapse | ko04725 | 79 | 2.90E-21 |
| 胃酸分泌 Gastric acid secretion | ko04971 | 69 | 2.05E-21 |
| 醛固酮合成与分泌 Aldosterone synthesis and secretion | ko04925 | 74 | 1.94E-21 |
| cAMP信号通路 cAMP signaling pathway | ko04024 | 113 | 1.73E-21 |
| 昼夜节律调节 Circadian entrainment | ko04713 | 75 | 2.93E-23 |
| 胰岛素分泌 Insulin secretion | ko04911 | 78 | 2.53E-23 |
| ErbB信号通路 ErbB signaling pathway | ko04012 | 80 | 2.43E-25 |
表6
意大利蜜蜂工蜂中肠下调miRNA的靶mRNA显著富集的前15位通路"
| 通路 Pathway | 通路ID Pathway ID | 靶mRNA富集数 Number of target mRNAs | P值 P value |
|---|---|---|---|
| Hippo信号通路 Hippo signaling pathway | ko04390 | 134 | 6.20E-17 |
| 醛固酮合成与分泌 Aldosterone synthesis and secretion | ko04925 | 78 | 1.79E-17 |
| 胆碱能突触 Cholinergic synapse | ko04725 | 84 | 1.71E-17 |
| 卵母细胞减数分裂 Oocyte meiosis | ko04114 | 103 | 8.04E-19 |
| 昼夜节律调节 Circadian entrainment | ko04713 | 79 | 3.12E-19 |
| 胃酸分泌 Gastric acid secretion | ko04971 | 75 | 1.66E-19 |
| 神经营养因子信号通路 Neurotrophin signaling pathway | ko04722 | 99 | 1.20E-19 |
| 胰岛素分泌 Insulin secretion | ko04911 | 83 | 9.94E-20 |
| 促性腺激素释放激素信号通路 GnRH signaling pathway | ko04912 | 91 | 9.12E-20 |
| 长时程增强 Long-term potentiation | ko04720 | 82 | 4.72E-20 |
| 钙信号通道 Calcium signaling pathway | ko04020 | 101 | 3.09E-22 |
| ErbB信号通路 ErbB signaling pathway | ko04012 | 87 | 3.11E-23 |
| cAMP信号通路 cAMP signaling pathway | ko04024 | 134 | 1.19E-23 |
| 炎症介质对TRP通道的调节Inflammatory mediator regulation of TRP channels | ko04750 | 97 | 9.4E-26 |
| 黑色素生成 Melanogenesis | ko04916 | 117 | 9.23E-28 |
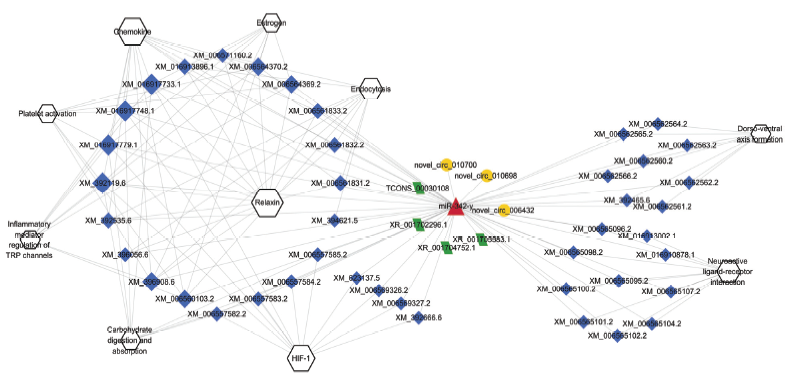
图4
意蜂工蜂中肠的miR-342-y靶向结合的10条信号通路相关DElncRNA、DEcircRNA、DEmRNA关系网络 六边形代表信号通路 Hexagons indicate signaling pathways;红色三角形代表miRNA Red triangle indicates miRNA;黄色圆形代表DEcircRNA Yellow circles indicate DEcircRNAs;绿色平行四边形代表DElncRNA Green parallelograms indicate DElncRNAs;蓝色菱形代表mRNAs Blue diamonds indicate mRNAs;六边形和菱形的大小代表连接mRNA和信号通路的数量 The size of hexagon and diamond indicates the number of connected DEmRNAs or signaling pathways"
| [1] |
刘朋飞, 吴杰, 李海燕, 林素文. 中国农业蜜蜂授粉的经济价值评估. 中国农业科学, 2011,44(24):5117-5123.
doi: 10.3864/j.issn.0578-1752.2011.24.018 |
|
LIU P F, WU J, LI H Y, LIN S W. Economic values of bee pollination to China’s agriculture. Scientia Agricultura Sinica, 2011,44(24):5117-5123. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2011.24.018 |
|
| [2] | 梁勤, 陈大福. 蜜蜂保护学. 北京: 中国农业出版社, 2009. |
| LIANG Q, CHEN D F. Bee Conservation. Beijing: China Agriculture Press, 2009. (in Chinese) | |
| [3] |
GUO R, WANG S M, XUE R Y, CAO G L, HU X L, HUANG M L, ZHANG Y Q, LU Y H, ZHU L Y, CHEN F, LIANG Z, KUANG S L, GONG C L. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Applied Microbiology and Biotechnology, 2015,99(12):5175-5187.
doi: 10.1007/s00253-015-6634-x pmid: 25957492 |
| [4] |
BARTEL D P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 2004,116(2):281-297.
doi: 10.1016/s0092-8674(04)00045-5 pmid: 14744438 |
| [5] |
PILLAI R S, BHATTACHARYYA S N, ARTUS C G, ZOLLER T, COUGOT N, BASYUK E, BERTRAND E, FILIPOWICZ W. Inhibition of translational initiation by let-7 microRNA in human cells. Science, 2005,309(5740):1573-1576.
doi: 10.1126/science.1115079 pmid: 16081698 |
| [6] |
BRENNECKE J, HIPFNER D R, STARK A, RUSSELL R B, COHEN S M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell, 2003,113(1):25-36.
doi: 10.1016/s0092-8674(03)00231-9 pmid: 12679032 |
| [7] |
XU P, VERNOOY S Y, GUO M, HAY B A. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Current Biology, 2003,13(9):790-795.
doi: 10.1016/S0960-9822(03)00250-1 |
| [8] |
LEE R C, FEINBAUM R L, AMBROS V. TheC.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 1993,75(5):843-854.
doi: 10.1016/0092-8674(93)90529-y pmid: 8252621 |
| [9] | 熊翠玲, 杜宇, 陈大福, 郑燕珍, 付中民, 王海鹏, 耿四海, 陈华枝, 周丁丁, 吴素珍, 石彩云, 郭睿. 意大利蜜蜂幼虫肠道的miRNAs的生物信息学预测及分析. 应用昆虫学报, 2018,55(6):1023-1033. |
| XIONG C L, DU Y, CHEN D F, ZHENG Y Z, FU Z M, WANG H P, GENG S H, CHEN H Z, ZHOU D D, WU S Z, SHI C Y, GUO R. Bioinformatic prediction and analysis of miRNAs in the Apis mellifera ligustica larval gut. Chinese Journal of Applied Entomology, 2018, 55(6):1023-1033. (in Chinese) | |
| [10] |
GUO Y, LIU H, YANG Z, CHEN J, SUN Y, REN X. Identification and characterization of miRNAome in tobacco (Nicotiana tabacum) by deep sequencing combined with microarray. Gene, 2012,501(1):24-32.
doi: 10.1016/j.gene.2012.04.002 |
| [11] |
郭睿, 王海朋, 陈华枝, 熊翠玲, 郑燕珍, 付中民, 赵红霞, 陈大福. 蜜蜂球囊菌的microRNA鉴定及其调控网络分析. 微生物学报, 2018,58(6):1077-1089.
doi: 10.13343/j.cnki.wsxb.20170535 |
|
GUO R, WANG H P, CHEN H Z, XIONG C L, ZHENG Y Z, FU Z M, ZHAO H X, CHEN D F. Identification of Ascosphaera apis microRNAs and investigation of their regulation networks. Acta Microbiologica Sinica, 2018,58(6):1077-1089. (in Chinese)
doi: 10.13343/j.cnki.wsxb.20170535 |
|
| [12] |
ENRIGHT A, JOHN B, GAUL U, TUSCHL T, SANDER C, MARKS D. MicroRNA targets inDrosophila. Genome Biology, 2003,4(11):P8.
doi: 10.1186/gb-2003-4-11-p8 |
| [13] |
LIU F, PENG W, LI Z, LI W, LI L, PAN J, ZHANG S, MIAO Y, CHEN S, SU S. Next-generation small RNA sequencing for microRNAs profiling inApis mellifera: Comparison between nurses and foragers. Insect Molecular Biology, 2012,21(3):297-303.
doi: 10.1111/j.1365-2583.2012.01135.x |
| [14] |
SHI Y Y, ZHENG H J, PAN Q Z, WANG Z L, ZENG Z J. Differentially expressed microRNAs between queen and worker larvae of the honey bee (Apis mellifera). Apidologie, 2015,46:35-45.
doi: 10.1007/s13592-014-0299-9 |
| [15] |
LOURENÇO A P, GUIDUGLI-LAZZARINI K R, FREITAS F C P, BITONDI M M G, SIMÕES Z L P. Bacterial infection activates the immune system response and dysregulates microRNA expression in honey bees. Insect Biochemistry and Molecular Biology, 2013,43(5):474-482.
doi: 10.1016/j.ibmb.2013.03.001 |
| [16] |
SALMENA L, POLISENO L, TAY Y, KATS L, PANDOLFI P P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell, 2011,146(3):353-358.
doi: 10.1016/j.cell.2011.07.014 |
| [17] |
TAY Y, RINN J, PANDOLFI P P. The multilayered complexity of ceRNA crosstalk and competition. Nature, 2014,505(7483):344-352.
doi: 10.1038/nature12986 |
| [18] |
WANG K, LIU C Y, ZHOU L Y, WANG J X, WANG M, ZHAO B, ZHAO W K, XU S J, FAN L H, ZHANG X J, FENG C, WANG C Q, ZHAO Y F, LI P F. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nature Communications, 2015,6:6779.
doi: 10.1038/ncomms7779 pmid: 25858075 |
| [19] |
郭睿, 杜宇, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 王海朋, 陈华枝, 耿四海, 周丁丁, 石彩云, 赵红霞, 陈大福. 意大利蜜蜂幼虫肠道发育过程中的差异表达microRNA及其调控网络. 中国农业科学, 2018,51(21):4197-4209.
doi: 10.3864/j.issn.0578-1752.2018.21.018 |
|
GUO R, DU Y, XIONG C L, ZHENG Y Z, FU Z M, XU G J, WANG H P, CHEN H Z, GENG S H, ZHOU D D, SHI C Y, ZHAO H X, CHEN D F. Differentially expressed microRNA and their regulation networks during the developmental process of Apis mellifera ligustica larval gut. Scientia Agricultura Sinica, 2018,51(21):4197-4209. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2018.21.018 |
|
| [20] |
郭睿, 耿四海, 熊翠玲, 郑燕珍, 付中民, 王海鹏, 杜宇, 童新宇, 赵红霞, 陈大福. 意大利蜜蜂工蜂中肠发育过程中长链非编码RNA的差异表达分析. 中国农业科学, 2018,51(18):3600-3613.
doi: 10.3864/j.issn.0578-1752.2018.18.016 |
|
GUO R, GENG S H, XIONG C L, ZHENG Y Z, FU Z M, WANG H P, DU Y, TONG X Y, ZHAO H X, CHEN D F. Differential expression analysis of long non-coding RNAs during the developmental process of Apis mellifera ligustica worker’s midgut. Scientia Agricultura Sinica, 2018,51(18):3600-3613. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2018.18.016 |
|
| [21] |
郭睿, 陈华枝, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 杜宇, 王海朋, 耿四海, 周丁丁, 刘思亚, 陈大福. 意大利蜜蜂工蜂中肠发育过程中的差异表达环状RNA及其调控网络分析. 中国农业科学, 2018,51(23):4575-4590.
doi: 10.3864/j.issn.0578-1752.2018.23.015 |
|
GUO R, CHEN H Z, XIONG C L, ZHENG Y Z, FU Z M, XU G J, DU Y, WANG H P, GENG S H, ZHOU D D, LIU S Y, CHEN D F. Analysis of differentially expressed circular RNAs and their regulation networks during the developmental process of Apis mellifera ligustica worker’s midgut. Scientia Agricultura Sinica, 2018,51(23):4575-4590. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2018.23.015 |
|
| [22] |
杜宇, 周丁丁, 万洁琦, 卢家轩, 范小雪, 范元婵, 陈恒, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 陈大福, 郭睿. 意大利蜜蜂工蜂中肠发育过程中的差异基因表达谱及调控网络. 中国农业科学, 2020,53(1):201-212.
doi: 10.3864/j.issn.0578-1752.2020.01.019 |
|
DU Y, ZHOU D D, WAN J Q, LU J X, FAN X X, FAN Y C, CHEN H, XIONG C L, ZHENG Y Z, FU Z M, XU G J, CHEN D F, GUO R. Profiling and regulation network of differentially expressed genes during the development process of Apis mellifera ligustica worker’s midgut. Scientia Agricultura Sinica, 2020,53(1):201-212. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2020.01.019 |
|
| [23] | 杜宇, 童新宇, 周丁丁, 陈大福, 熊翠玲, 郑燕珍, 徐国钧, 王海朋, 陈华枝, 郭意龙, 隆琦, 郭睿. 中华蜜蜂幼虫肠道响应球囊菌胁迫的microRNA应答分析. 微生物学报, 2019,59(9):1747-1764. |
| DU Y, TONG X Y, ZHOU D D, CHEN D F, XIONG C L, ZHENG Y Z, XU G J, WANG H P, CHEN H Z, GUO Y L, LONG Q, GUO R. MicroRNA responses in the larval gut of Apis cerana cerana to Ascosphaera apis stress. Acta Microbiologica Sinica, 2019,59(9):1747-1764. (in Chinese) | |
| [24] |
郭睿, 杜宇, 童新宇, 熊翠玲, 郑燕珍, 徐国钧, 王海朋, 耿四海, 周丁丁, 郭意龙, 吴素珍, 陈大福. 意大利蜜蜂幼虫肠道在球囊菌侵染前期的差异表达microRNA及其调控网络. 中国农业科学, 2019,52(1):166-180.
doi: 10.3864/j.issn.0578-1752.2019.01.015 |
|
GUO R, DU Y, TONG X Y, XIONG C L, ZHENG Y Z, XU G J, WANG H P, GENG S H, ZHOU D D, GUO Y L, WU S Z, CHEN D F. Differentially expressed microRNAs and their regulation networks in Apis mellifera ligustica larval gut during the early stage of Ascosphaera apis infection. Scientia Agricultura Sinica, 2019,52(1):166-180. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2019.01.015 |
|
| [25] |
VALLET-GELY I, LEMAITRE B, BOCCARD F. Bacterial strategies to overcome insect defences. Nature Reviews Microbiology, 2008,6(4):302-313.
doi: 10.1038/nrmicro1870 pmid: 18327270 |
| [26] |
RAES H, VERBEKE M, MEULEMANS W, COSTER W D. Organisation and ultrastructure of the regenerative crypts in the midgut of the adult worker honeybee (Apis mellifera L.). Tissue and Cell, 1994,26(2):231-238.
doi: 10.1016/0040-8166(94)90098-1 pmid: 18621268 |
| [27] |
GENG L L, CUI H J, DAI P L, LANG Z H, SHU C L, ZHOU T, SONG F P, ZHANG J. The influence of Bt-transgenic maize pollen on the bacterial diversity in the midgut ofApis mellifera ligustica. Apidologie, 2013,44(2):198-208.
doi: 10.1007/s13592-012-0171-8 |
| [28] |
CHEN D F, CHEN H Z, DU Y, ZHOU D D, GENG S H, WANG H P, WAN J Q, XIONG C L, ZHENG Y Z, GUO R. Genome-wide identification of long non-coding RNAs and their regulatory networks involved inApis mellifera ligustica response to Nosema ceranae infection. Insects, 2019,10(8):245.
doi: 10.3390/insects10080245 |
| [29] |
CHEN D F, DU Y, CHEN H Z, FAN Y C, FAN X X, ZHU Z W, WANG J, XIONG C L, ZHENG Y Z, HOU C S, DIAO Q Y, GUO R. Comparative identification of microRNAs inApis cerana cerana workers’ midguts in response to Nosema ceranae invasion. Insects, 2019,10(9):258.
doi: 10.3390/insects10090258 |
| [30] |
GUO R, CHEN D F, CHEN H Z, XIONG C L, ZHENG Y Z, HOU C S, DU Y, GENG S H, WANG H P, ZHOU D D, GUO Y L. Genome-wide identification of circular RNAs in fungal parasiteNosema ceranae. Current Microbiology, 2018,75(12):1655-1660.
doi: 10.1007/s00284-018-1576-z pmid: 30269253 |
| [31] |
付中民, 陈华枝, 刘思亚, 祝智威, 范小雪, 范元婵, 万洁琦, 张璐, 熊翠玲, 徐国钧, 陈大福, 郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答. 中国农业科学, 2019,52(17):3069-3082.
doi: 10.3864/j.issn.0578-1752.2019.17.014 |
|
FU Z M, CHEN H Z, LIU S Y, ZHU Z W, FAN X X, FAN Y C, WAN J Q, ZHANG L, XIONG C L, XU G J, CHEN D F, GUO R. Immune responses of Apis mellifera ligustia to Nosema ceranae stress. Scientia Agricultura Sinica, 2019,52(17):3069-3082. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2019.17.014 |
|
| [32] | 房宇. 蜜蜂工蜂和雄蜂胚胎期发育蛋白质组及磷酸化蛋白质组研究[D]. 北京: 中国农业科学院, 2017. |
| FANG Y. Unraveling the molecular underpinnings of embryonic development of honeybee worker and drone (Apis mellifera ligustica) using proteomics and phosphoproteomics[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. (in Chinese) | |
| [33] | 王超, 张文翔, 殷梦昕, 张雷. Hippo信号调控果蝇中肠稳态维持的机制研究. 中国细胞生物学学报, 2015,37(5):599-603. |
| WANG C, ZHANG W X, YIN M X, ZHANG L. The study on the regulatory mechanisms of Drosophila midgut homeostasis by Hippo signaling. Chinese Journal of Cell Biology, 2015,37(5):599-603. (in Chinese) | |
| [34] |
OHLSTEIN B, SPRADLING A. MultipotentDrosophila intestinal stem cells specify daughter cell fates by differential Notch signaling. Science, 2007,315(5814):988-992.
doi: 10.1126/science.1136606 pmid: 17303754 |
| [35] |
HARTENSTEIN A Y, RUGENDORFF A, TEPASS U, HARTENSTEIN V. The function of the neurogenic genes during epithelial development in theDrosophila embryo. Development, 1992,116(4):1203-1220.
pmid: 1295737 |
| [36] |
MICCHELLI C A, PERRIMON N. Evidence that stem cells reside in the adultDrosophila midgut epithelium. Nature, 2006,439(7075):475-479.
doi: 10.1038/nature04371 pmid: 16340959 |
| [37] |
OHLSTEIN B, SPRADLING A. The adultDrosophila posterior midgut is maintained by pluripotent stem cells. Nature, 2006,439(7075):470-474.
doi: 10.1038/nature04333 pmid: 16340960 |
| [38] |
TIAN A, BENCHABANE H, WANG Z, AHMED Y. Regulation of stem cell proliferation and cell fate specification by Wingless/Wnt signaling gradients enriched at adult intestinal compartment boundaries. PLoS Genetics, 2016,12(2):e1005822.
doi: 10.1371/journal.pgen.1005822 pmid: 26845150 |
| [39] |
REN F, WANG B, YUE T, YUN E Y, IP Y T, JIANG J. Hippo signaling regulatesDrosophila intestine stem cell proliferation through multiple pathways. Proceedings of the National Academy of Sciences of the United States of America, 2010,107(49):21064-21069.
doi: 10.1073/pnas.1012759107 pmid: 21078993 |
| [40] | 宋茜. FoxO基因在昆虫蜕皮中的功能研究[D]. 济南: 山东大学, 2013. |
| SONG Q. Functional research of FoxO gene in insect molt in Helicoverpa armigera[D]. Ji’nan: Shandong University, 2013 .(in Chinese) | |
| [41] |
EVANS J D, ARMSTRONG T N. Antagonistic interactions between honey bee bacterial symbionts and implications for disease. BMC Ecology, 2006,6:4.
doi: 10.1186/1472-6785-6-4 pmid: 16551367 |
| [42] |
ARBOUZOVA N I, ZEIDLER M P. JAK/STAT signalling inDrosophila: Insights into conserved regulatory and cellular functions. Development, 2006,133(14):2605-2616.
doi: 10.1242/dev.02411 pmid: 16794031 |
| [43] |
KIM T, KIM Y J. Overview of innate immunity inDrosophila. Journal of Biochemistry and Molecular Biology, 2005,38(2):121-127.
doi: 10.5483/bmbrep.2005.38.2.121 pmid: 15826489 |
| [44] |
SCHUMACHER M A, FENG R, AIHARA E, ENGEVIK A C, MONTROSE M H, OTTEMANN K M, ZAVROS Y. Helicobacter pylori-induced Sonic Hedgehog expression is regulated by NF-κB pathway activation: The use of a novel in vitro model to study epithelial response to infection. Helicobacter, 2015,20(1):19-28.
doi: 10.1111/hel.12152 pmid: 25495001 |
| [45] |
SEGER R, KREBS E G. The MAPK signaling cascade. FASEB Journal, 1995,9(9):726-735.
pmid: 7601337 |
| [46] |
CONSTANT S L, DONG C, YANG D D, WYSK M, DAVIS R J, FLAVELL R A. JNK1 is required for T cell-mediated immunity againstLeishmania major infection. Journal of Immunology, 2000,165(5):2671-2676.
doi: 10.4049/jimmunol.165.5.2671 |
| [47] |
JANSSENS V, GORIS J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. The Biochemical Journal, 2001,353(3):417-439.
doi: 10.1042/bj3530417 |
| [48] | 牛陵川, 李长清. cAMP-PKA信号通路与轴突再生. 国际神经病学神经外科学杂志, 2007,34(3):290-293. |
| NIU L C, LI C Q. Effect of cAMP-PKA signal pathway on axon regeneration. Journal of International Neurology and Neurosurgery, 2007,34(3):290-293. (in Chinese) | |
| [49] |
ZHOU R S, ZHANG E X, SUN Q F, YE Z J, LIU J W, ZHOU D H, TANG Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer, 2019,19(1):779.
doi: 10.1186/s12885-019-5983-8 pmid: 31391008 |
| [50] | 裘智勇, 赵巧玲, 刘挺, 覃光星, 沈兴家, 郭锡杰. 两个对浓核病毒(镇江株)具感性差异的家蚕品种的血液和围食膜蛋白的比较. 基因组学与应用生物学, 2011,30(3):274-281. |
| QIU Z Y, ZHAO Q L, LIU T, QIN X G, SHEN X J, GUO X J. Comparison of proteins derived from hemolymph and peritrophic membrane of two silkworm strains with different susceptibility to Bombyx mori densovirus (Zhenjiang strain). Genomics and Applied Biology, 2011,30(3):274-281. (in Chinese) | |
| [51] | 钟晓武. 家蚕围食膜的蛋白质组及几丁质去乙酰化酶的功能研究[D]. 重庆: 西南大学, 2012. |
| ZHONG X W. Proteomic analysis on peritrophic membrane and functional characterization of chitin deacetylase in silkworm, Bombyx mori[D]. Chongqing: Southwest University, 2012. (in Chinese) |
| [1] | 王荣华,孟丽峰,冯毛,房宇,魏俏红,马贝贝,钟未来,李建科. 蜂王浆高产蜜蜂与意大利蜜蜂哺育蜂唾液腺蛋白质组分析[J]. 中国农业科学, 2022, 55(13): 2667-2684. |
| [2] | 冯睿蓉,付中民,杜宇,张文德,范小雪,王海朋,万洁琦,周紫彧,康育欣,陈大福,郭睿,史培颖. 中华蜜蜂幼虫肠道中微小RNA的鉴定及分析[J]. 中国农业科学, 2022, 55(1): 208-218. |
| [3] | 杜宇,范小雪,蒋海宾,王杰,冯睿蓉,张文德,余岢骏,隆琦,蔡宗兵,熊翠玲,郑燕珍,陈大福,付中民,徐国钧,郭睿. 微小RNA介导意大利蜜蜂工蜂对东方蜜蜂微孢子虫的跨界调控[J]. 中国农业科学, 2021, 54(8): 1805-1820. |
| [4] | 陈华枝,王杰,祝智威,蒋海宾,范元婵,范小雪,万洁琦,卢家轩,郑燕珍,付中民,徐国钧,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中长链非编码RNA的比较及潜在功能分析[J]. 中国农业科学, 2021, 54(2): 435-448. |
| [5] | 王继卿,郝志云,沈继源,柯娜,黄兆春,梁维炜,罗玉柱,胡江,刘秀,李少斌. 小尾寒羊泌乳性状重要lncRNAs的筛选、鉴定及功能分析[J]. 中国农业科学, 2021, 54(14): 3113-3123. |
| [6] | 周丁丁, 范元婵, 王杰, 蒋海宾, 祝智威, 范小雪, 陈华枝, 杜宇, 周紫彧, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 蜜蜂球囊菌中长链非编码RNA的调控作用[J]. 中国农业科学, 2021, 54(1): 224-238. |
| [7] | 贾海燕,宋丽云,徐翔,解屹,张超群,刘天波,赵存孝,申莉莉,王杰,李莹,王凤龙,杨金广. 不同温度下TMV侵染枯斑三生烟的LncRNA差异表达[J]. 中国农业科学, 2020, 53(7): 1381-1396. |
| [8] | 高艳,朱雅楠,李秋方,苏松坤,聂红毅. 转录组学分析意大利蜜蜂脑部哺育行为相关基因[J]. 中国农业科学, 2020, 53(19): 4092-4102. |
| [9] | 陈华枝,祝智威,蒋海宾,王杰,范元婵,范小雪,万洁琦,卢家轩,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中微小RNA及其靶mRNA的比较分析[J]. 中国农业科学, 2020, 53(17): 3606-3619. |
| [10] | 耿四海,石彩云,范小雪,王杰,祝智威,蒋海宾,范元婵,陈华枝,杜宇,王心蕊,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 微小RNA介导东方蜜蜂微孢子虫侵染意大利蜜蜂工蜂的分子机制[J]. 中国农业科学, 2020, 53(15): 3187-3204. |
| [11] | 周丁丁,史小玉,王杰,范元婵,祝智威,蒋海宾,范小雪,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 东方蜜蜂微孢子虫孢子中长链非编码RNA的竞争性内源RNA调控网络及潜在功能[J]. 中国农业科学, 2020, 53(10): 2122-2136. |
| [12] | 杜宇,周丁丁,万洁琦,卢家轩,范小雪,范元婵,陈恒,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 意大利蜜蜂工蜂中肠发育过程中的差异基因 表达谱及调控网络[J]. 中国农业科学, 2020, 53(1): 201-212. |
| [13] | 付中民,陈华枝,刘思亚,祝智威,范小雪,范元婵,万洁琦,张璐,熊翠玲,徐国钧,陈大福,郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答[J]. 中国农业科学, 2019, 52(17): 3069-3082. |
| [14] | 于静,张卫星,马兰婷,胥保华. 饲粮α-亚麻酸水平对意大利蜜蜂工蜂幼虫生理机能的影响[J]. 中国农业科学, 2019, 52(13): 2368-2378. |
| [15] | 郭睿,杜宇,童新宇,熊翠玲,郑燕珍,徐国钧,王海朋,耿四海,周丁丁,郭意龙,吴素珍,陈大福. 意大利蜜蜂幼虫肠道在球囊菌侵染前期的 差异表达microRNA及其调控网络[J]. 中国农业科学, 2019, 52(1): 166-180. |
|
||