













中国农业科学 ›› 2021, Vol. 54 ›› Issue (8): 1805-1820.doi: 10.3864/j.issn.0578-1752.2021.08.019
• 畜牧·兽医·资源昆虫 • 上一篇
杜宇1( ),范小雪1(
),范小雪1( ),蒋海宾1,王杰1,冯睿蓉1,张文德1,余岢骏1,隆琦1,蔡宗兵1,熊翠玲1,郑燕珍1,2,陈大福1,2,付中民1,2,徐国钧1,2,郭睿1,2(
),蒋海宾1,王杰1,冯睿蓉1,张文德1,余岢骏1,隆琦1,蔡宗兵1,熊翠玲1,郑燕珍1,2,陈大福1,2,付中民1,2,徐国钧1,2,郭睿1,2( )
)
收稿日期:2020-06-02
接受日期:2020-06-22
出版日期:2021-04-16
发布日期:2021-04-25
通讯作者:
郭睿
作者简介:杜宇,E-mail: 基金资助:
DU Yu1( ),FAN XiaoXue1(
),FAN XiaoXue1( ),JIANG HaiBin1,WANG Jie1,FENG RuiRong1,ZHANG WenDe1,YU KeJun1,LONG Qi1,CAI ZongBing1,XIONG CuiLing1,ZHENG YanZhen1,2,CHEN DaFu1,2,FU ZhongMin1,2,XU GuoJun1,2,GUO Rui1,2(
),JIANG HaiBin1,WANG Jie1,FENG RuiRong1,ZHANG WenDe1,YU KeJun1,LONG Qi1,CAI ZongBing1,XIONG CuiLing1,ZHENG YanZhen1,2,CHEN DaFu1,2,FU ZhongMin1,2,XU GuoJun1,2,GUO Rui1,2( )
)
Received:2020-06-02
Accepted:2020-06-22
Online:2021-04-16
Published:2021-04-25
Contact:
Rui GUO
摘要:
【目的】东方蜜蜂微孢子虫(Nosema ceranae)感染意大利蜜蜂(Apis mellifera ligustica,简称意蜂)导致蜜蜂微孢子虫病。本研究结合前期已获得的miRNA和mRNA组学数据,通过生物信息学方法对意蜂工蜂中肠的差异表达miRNA(differentially expressed miRNA,DEmiRNA)靶向结合的东方蜜蜂微孢子虫的mRNA和差异表达mRNA(DEmRNA)进行预测、数据库注释和调控网络分析,以期在组学水平解析miRNA介导意蜂工蜂对东方蜜蜂微孢子虫的跨界调控机制。【方法】通过比较东方蜜蜂微孢子虫侵染7 d和10 d的意蜂工蜂中肠(AmT1、AmT2)和未受侵染的工蜂中肠(AmCK1、AmCK2)的miRNA组学数据筛选出宿主的显著性DEmiRNA,通过比较侵染意蜂工蜂中肠的东方蜜蜂微孢子虫(NcT1、NcT2)和东方蜜蜂微孢子虫纯净孢子(NcCK)的mRNA数据筛选出病原的DEmRNA。利用TargetFinder软件预测宿主显著性DEmiRNA靶向结合的病原mRNA和DEmRNA。利用相关生物信息学工具对上述靶DEmRNA进行GO和KEGG数据库注释。结合前期研究结果筛选出孢壁蛋白、极管蛋白、蓖麻毒素B凝集素、ABC转运蛋白、ATP/ADP移位酶和糖酵解/糖异生途径等毒力因子和能量代谢通路相关的病原DEmRNA及与其存在靶向结合关系的宿主显著性DEmiRNA,并构建和分析二者的调控网络。【结果】AmCK1 vs AmT1比较组中宿主的48条显著上调miRNA和36条显著下调miRNA分别靶向病原的1 345和1 046条mRNA;进一步分析发现,宿主的47条显著上调miRNA和34条显著下调miRNA可分别靶向NcCK vs NcT1比较组中病原的584条显著下调mRNA和265条显著上调mRNA,它们可分别注释到19和22个功能条目以及66和64条通路。AmCK2 vs AmT2比较组中宿主的56条显著上调miRNA和51条显著下调miRNA分别靶向病原的1 260和1 317条mRNA;进一步分析发现,宿主的52条显著上调miRNA和49条显著下调miRNA可分别靶向NcCK vs NcT2比较组中病原的587条显著下调mRNA和336条显著上调mRNA,它们可分别注释到20和23个功能条目以及64和65条通路。AmCK1 vs AmT1和AmCK2 vs AmT2比较组的8条共同显著上调miRNA和1条共同显著下调miRNA分别靶向NcCK vs NcT1和NcCK vs NcT2比较组中的144条共同显著下调和10条共同显著上调mRNA,可分别注释到18和13个功能条目以及38和7条通路。此外,AmCK1 vs AmT1和AmCK2 vs AmT2比较组中宿主的显著上调miRNA可靶向结合NcCK vs NcT1和NcCK vs NcT2比较组中与RNAi途径,孢壁蛋白和蓖麻毒素B凝集素等毒力因子,糖酵解/糖异生途径以及MAPK信号通路相关的病原下调表达mRNA。【结论】在东方蜜蜂微孢子虫的侵染过程中,意蜂工蜂中肠的DEmiRNA与病原的DEmRNA之间存在复杂的靶向结合关系以及潜在的跨界调控关系;宿主的DEmiRNA可能通过抑制或降解病原的RNAi途径、毒力因子、糖酵解/糖异生通路、ATP/ADP移位酶、ABC转运蛋白及MAPK信号通路相关靶DEmRNA影响病原的侵染和增殖。
杜宇,范小雪,蒋海宾,王杰,冯睿蓉,张文德,余岢骏,隆琦,蔡宗兵,熊翠玲,郑燕珍,陈大福,付中民,徐国钧,郭睿. 微小RNA介导意大利蜜蜂工蜂对东方蜜蜂微孢子虫的跨界调控[J]. 中国农业科学, 2021, 54(8): 1805-1820.
DU Yu,FAN XiaoXue,JIANG HaiBin,WANG Jie,FENG RuiRong,ZHANG WenDe,YU KeJun,LONG Qi,CAI ZongBing,XIONG CuiLing,ZHENG YanZhen,CHEN DaFu,FU ZhongMin,XU GuoJun,GUO Rui. MicroRNA-Mediated Cross-Kingdom Regulation of Apis mellifera ligustica Worker to Nosema ceranae[J]. Scientia Agricultura Sinica, 2021, 54(8): 1805-1820.

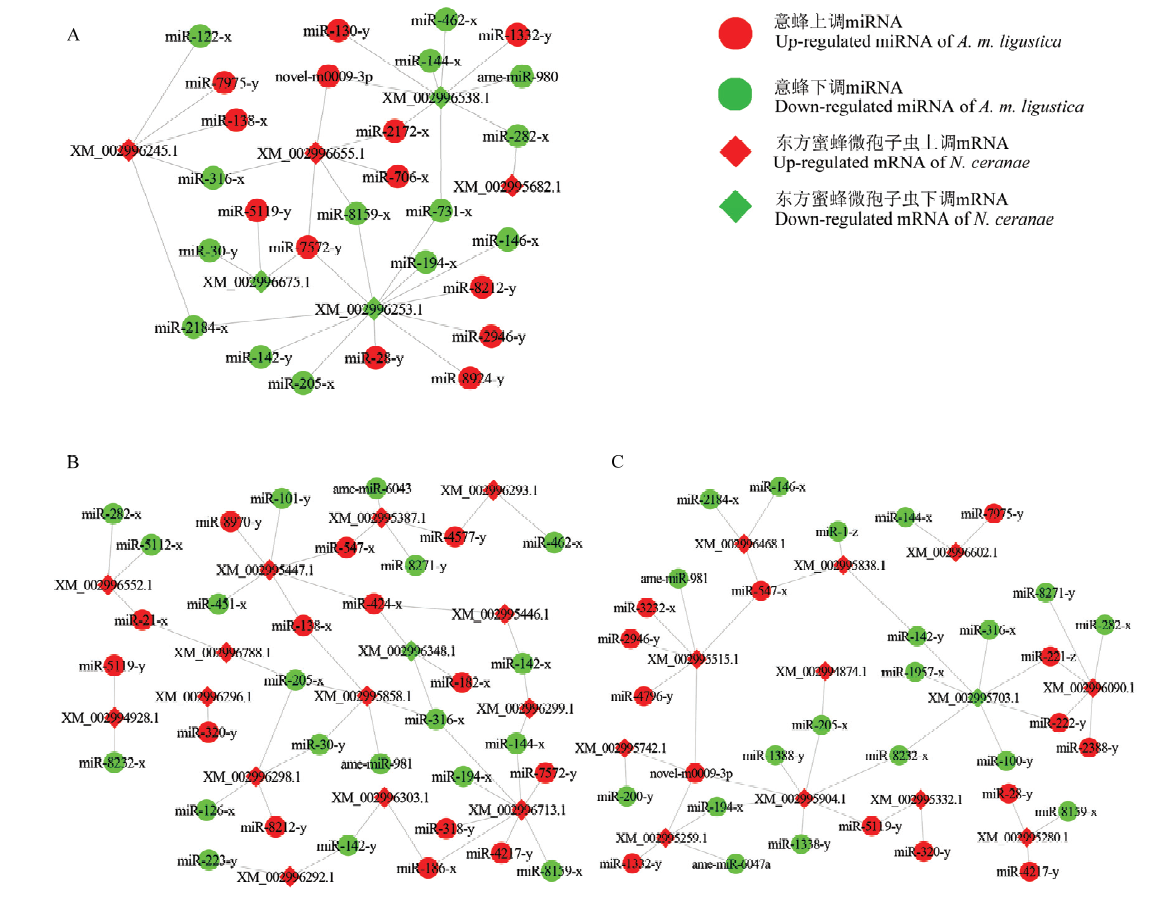
图1
意蜂工蜂中肠显著性DEmiRNA靶向东方蜜蜂微孢子虫显著性DEmRNA的调控网络 A:AmCK1 vs AmT1中显著下调miRNA及其靶向NcCK vs NcT1中显著上调mRNA的调控网络Regulatory network of significantly down-regulated miRNAs in AmCK1 vs AmT1 and their target significantly up-regulated mRNAs in NcCK vs NcT1;B:AmCK1 vs AmT1中显著上调miRNA靶向NcCK vs NcT1中显著下调mRNA的调控网络Regulatory network of significantly up-regulated miRNAs in AmCK1 vs AmT1 and their target significantly down-regulated mRNAs in NcCK vs NcT1;C:AmCK2 vs AmT2中显著下调miRNA靶向NcCK vs NcT2中显著上调mRNA的调控网络Regulatory network of significantly down-regulated miRNAs in AmCK2 vs AmT2 and their target significantly up-regulated mRNAs in NcCK vs NcT2;D:AmCK2 vs AmT2中显著上调miRNA靶向NcCK vs NcT2中显著下调mRNA的调控网络Regulatory network of significantly up-regulated miRNAs in AmCK2 vs AmT2 and their target significantly down-regulated mRNAs in NcCK vs NcT2"
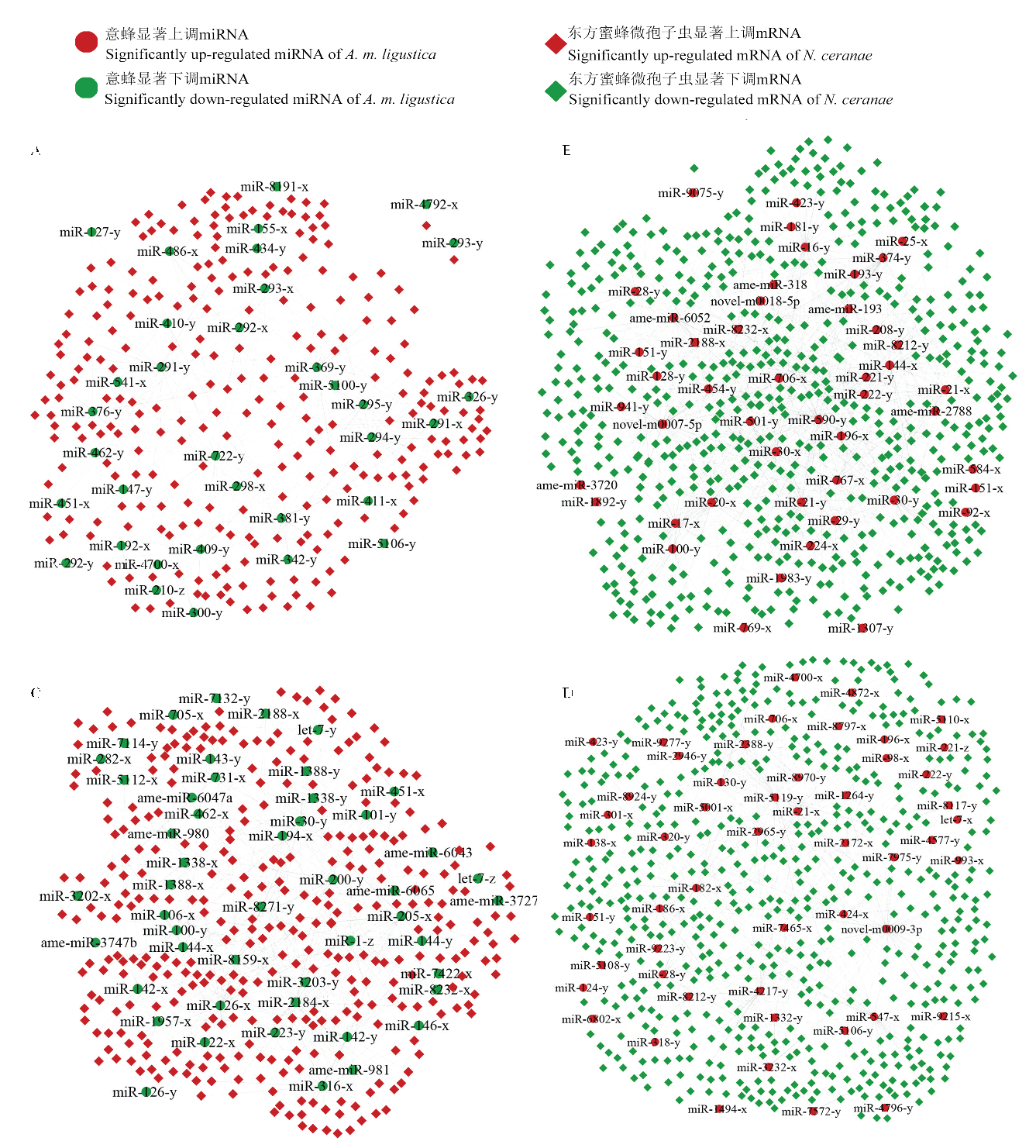
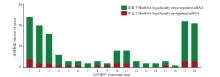
图3
AmCK1 vs AmT1和AmCK2 vs AmT2比较组的共同显著上调(下调)miRNA靶向NcCK vs NcT1和NcCK vs NcT2共同显著下调(上调)mRNA的GO数据库注释 1:代谢进程Metabolic process;2:细胞进程Cellular process;3:单一组织进程Single-organism process;4:应激反应Response to stimulus;5:信号Signaling;6:生物调节进程Biological regulation process;7:定位Localization;8:生物调节Biological regulation;9:细胞成分组织或生物合成Cellular component organization or biogenesis;10:细胞Cell;11:细胞组件Cell part;12:细胞器Organelle;13:细胞膜组件Cell membrane part;14:细胞膜Cell membrane;15:高分子复合物Macromolecular complex;16:细胞器组件Organelle part;17:催化活性Catalytic activity;18:结合Binding"

图4
AmCK1 vs AmT1比较组中显著性DEmiRNA及其靶向的NcCK vs NcT1比较组中毒力因子/侵染因子相关DEmRNA的调控网络 A:宿主的显著性DEmiRNA与病原的ABC转运蛋白、ATP/ADP转位酶相关DEmRNA的调控网络Regulatory network of host significant DEmiRNAs and pathogen DEmRNAs associated with ABC transporter and ATP/ADP translocase;B:宿主的显著性DEmiRNA与病原的蓖麻毒素B凝集素、孢壁蛋白和极管蛋白相关DEmRNA的调控网络Regulatory network of host significant DEmiRNAs and pathogen DEmRNAs associated with ricin B lectin, spore wall protein and polar tube protein;C:宿主的显著性DEmiRNA与病原的糖酵解/糖异生途径相关DEmRNA的调控网络Regulatory network of host significant DEmiRNAs and pathogen DEmRNAs associated with glycolysis/gluconeogenesis pathway"
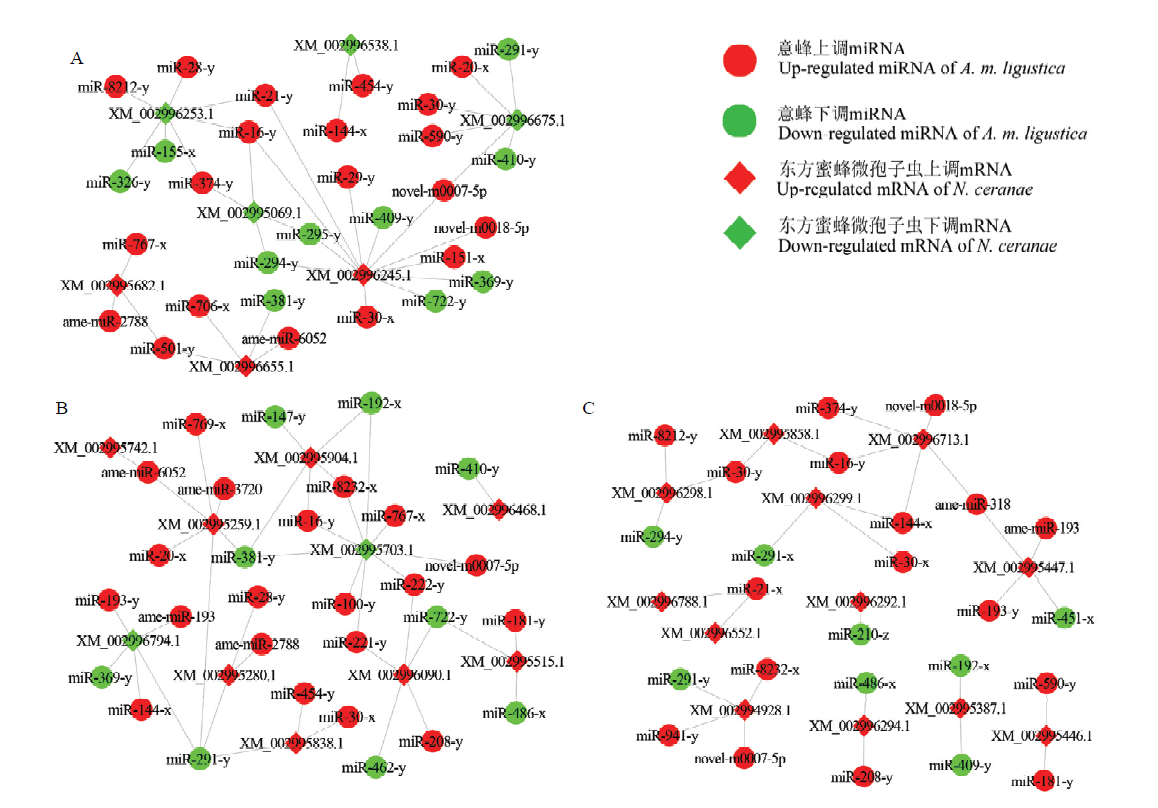
表1
AmCK1 vs AmT1中显著上调miRNA靶向NcCK vs NcT1中东方蜜蜂微孢子虫的毒力因子/侵染因子相关下调mRNA的信息概要"
| 差异表达miRNA DEmiRNA | 差异表达mRNA DEmRNA | 差异表达mRNA的log2FC log2FC of DEmRNA | 差异表达mRNA的P值 P value of DEmRNA | Nr数据库描述 Description in Nr database |
|---|---|---|---|---|
| ame-miR-6052 | XM_002996297.1 | -10.4546 | 0.0802 | 蓖麻毒素B凝集素 Ricin B lectin |
| miR-196-x | XM_002996297.1 | -10.4546 | 0.0802 | 蓖麻毒素B凝集素 Ricin B lectin |
| miR-16-y | XM_002995069.1 | -1.7206 | 2.6275E-25 | ABC转运蛋白 ABC transporter |
| miR-16-y | XM_002996253.1 | -3.6539 | 1.2369E-118 | ABC转运蛋白 ABC transporter |
| miR-20-x | XM_002996675.1 | -1.9081 | 5.2746E-22 | ABC转运蛋白 ABC transporter |
| miR-21-y | XM_002996253.1 | -3.6539 | 1.2369E-118 | ABC转运蛋白 ABC transporter |
| miR-28-y | XM_002996253.1 | -3.6539 | 1.2369E-118 | ABC转运蛋白 ABC transporter |
| miR-30-y | XM_002996675.1 | -1.9081 | 5.2746E-22 | ABC转运蛋白 ABC transporter |
| miR-374-y | XM_002995069.1 | -1.7206 | 2.6275E-25 | ABC转运蛋白 ABC transporter |
| miR-374-y | XM_002996253.1 | -3.6539 | 1.2369E-118 | ABC转运蛋白 ABC transporter |
| miR-590-y | XM_002996675.1 | -1.9081 | 5.2746E-22 | ABC转运蛋白 ABC transporter |
| miR-8212-y | XM_002996253.1 | -3.6539 | 1.2369E-118 | ABC转运蛋白 ABC transporter |
| novel-m0007-5p | XM_002996675.1 | -1.9081 | 5.2746E-22 | ABC转运蛋白 ABC transporter |
| miR-144-x | XM_002996538.1 | -3.6489 | 8.2034E-116 | ATP/ADP转位酶 ATP/ADP translocase |
| miR-454-y | XM_002996538.1 | -3.6489 | 8.2034E-116 | ATP/ADP转位酶 ATP/ADP translocase |
| ame-miR-193 | XM_002996794.1 | -1.1738 | 1.1394E-08 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-100-y | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-144-x | XM_002996794.1 | -1.1738 | 1.1394E-08 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-16-y | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-193-y | XM_002996794.1 | -1.1738 | 1.1394E-08 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-221-y | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-222-y | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-767-x | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-8232-x | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| novel-m0007-5p | XM_002995703.1 | -2.3880 | 1.2790E-52 | 糖酵解/糖异生 Glycolysis/gluconeogenesis |
| miR-16-y | XM_002996061.1 | -5.8896 | 2.4289E-21 | MAPK信号通路 MAPK signaling pathway |
| miR-29-y | XM_002995842.1 | -2.6200 | 1.1211E-28 | MAPK信号通路 MAPK signaling pathway |
| novel-m0007-5p | XM_002995842.1 | -2.6200 | 1.1211E-28 | MAPK信号通路 MAPK signaling pathway |
表2
靶向NcCK vs NcT1中病原毒力因子/侵染因子相关下调DEmRNA的AmCK1 vs AmT1中宿主显著上调miRNA的信息概要"
| 差异表达miRNA ID DEmiRNA ID | 差异表达miRNA的log2FC log2FC of DEmiRNA | 差异表达miRNA的P值 P value of DEmiRNA | AmCK1组中的TPM值 TPM in AmCK1 group | AmT1组中的TPM值 TPM in AmT1 group |
|---|---|---|---|---|
| ame-miR-6052 | 11.7271 | 0.0021 | 0.0010 | 3.3900 |
| miR-196-x | 2.3757 | 0.0012 | 3.2800 | 17.0233 |
| miR-16-y | 2.7925 | 0.0421 | 0.5100 | 3.5333 |
| miR-20-x | 1.9111 | 0.0500 | 2.2733 | 8.5500 |
| miR-21-y | 2.2041 | 0.0000 | 12.6300 | 58.1967 |
| miR-28-y | 1.6298 | 0.0072 | 7.7467 | 23.9733 |
| miR-30-y | 2.1421 | 0.0034 | 6.3900 | 28.2067 |
| miR-374-y | 4.7301 | 0.0009 | 0.2033 | 5.3967 |
| miR-590-y | 3.6286 | 0.0325 | 0.1900 | 2.3500 |
| miR-8212-y | 11.4683 | 0.0036 | 0.0010 | 2.8333 |
| novel-m0007-5p | 2.9010 | 0.0223 | 0.4900 | 3.6600 |
| miR-144-x | 4.2291 | 0.0307 | 0.3933 | 7.3767 |
| miR-454-y | 3.5531 | 0.0306 | 0.2033 | 2.3867 |
| ame-miR-193 | 3.1393 | 0.0039 | 0.9167 | 8.0767 |
| miR-100-y | 3.3495 | 0.0003 | 1.0367 | 10.5667 |
| miR-193-y | 2.8593 | 0.0049 | 0.8833 | 6.4100 |
| miR-221-y | 1.3376 | 0.0444 | 9.6667 | 24.4300 |
| miR-222-y | 1.8682 | 0.0000 | 100.1467 | 365.6167 |
| miR-767-x | 4.7167 | 0.0011 | 0.2033 | 5.3467 |
| miR-8232-x | 3.7159 | 0.0226 | 0.1900 | 2.4967 |
| miR-29-y | 1.5049 | 0.0014 | 29.6800 | 84.2333 |

图5
AmCK2 vs AmT2比较组中显著性DEmiRNA及其靶向的NcCK vs NcT2比较组中毒力因子/侵染因子相关DEmRNA的调控网络 A:宿主的显著性DEmiRNA与病原的ABC转运蛋白、ATP/ADP转位酶相关DEmRNA的调控网络Regulatory network of host significant DEmiRNAs and pathogen DEmRNAs associated with ABC transporter and ATP/ADP translocase;B:宿主的显著性DEmiRNA与病原的蓖麻毒素B凝集素、孢壁蛋白和极管蛋白相关DEmRNA的调控网络Regulatory network of host significant DEmiRNAs and pathogen DEmRNAs associated with ricin B lectin, spore wall protein and polar tube protein;C:宿主的显著性DEmiRNA与病原的糖酵解/糖异生途径相关DEmRNA的调控网络Regulatory network of host significant DEmiRNAs and pathogen DEmRNAs associated with glycolysis/gluconeogenesis pathway"
| [1] | GALLAI N, SALLES J M, SETTELE J, VAISSIERE B E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics, 2009,68(3):810-821. |
| [2] | WITTNER M, WEISS L M. The Microsporidia and Microsporidiosis. John Wiley & Sons, Inc., 1999. |
| [3] | MARTÍN-HERNÁNDEZ R, BARTOLOMÉ C, CHEJANOVSKY N, CONTE Y L, DALMON A, DUSSAUBAT C, GARCÍA-PALENCIA P, MEANA A, PINTO M A, SOROKER V, HIGES M. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environmental Microbiology, 2018,20(4):1302-1329. |
| [4] | MAYACK C, NATSOPOULOU M E, MCMAHON D P. Nosema ceranae alters a highly conserved hormonal stress pathway in honeybees. Insect Molecular Biology, 2015,24(6):662-670. |
| [5] | EVANS J D, HUANG Q. Interactions among host-parasite microRNAs during Nosema ceranae proliferation in Apis mellifera. Frontiers in Microbiology, 2018,9:698. |
| [6] |
BARTEL D P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 2004,116(2):281-297.
pmid: 14744438 |
| [7] |
ZHANG L, HOU D X, CHEN X, LI D H, ZHU L Y, ZHANG Y J, LI J, BIAN Z, LIANG X Y, CAI X, et al. Exogenous plant miR168a specifically targets mammalian LDLRAP1: Evidence of cross- kingdom regulation by microRNA. Cell Research, 2012,22(1):107-126.
pmid: 21931358 |
| [8] |
ZHU K, LIU M H, FU Z, ZHOU Z, KONG Y, LIANG H W, LIN Z G, LUO J, ZHENG H Q, WAN P, et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genetics, 2017,13(8):e1006946.
pmid: 28829772 |
| [9] |
CUI C L, WANG Y, LIU J N, ZHAO J, SUN P L, WANG S B. A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nature Communications, 2019,10(1):4298.
pmid: 31541102 |
| [10] | MAYORAL J G, HUSSAIN M, JOUBERT D A, ITURBE-ORMAETXE I, O’NEILL S L, ASGARI S. Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proceedings of the National Academy of Sciences of the United States of America, 2014,111(52):18721-18726. |
| [11] |
HINAS A, WRIGHT A J, HUNTER C P. SID-5 is an endosome- associated protein required for efficient systemic RNAi in C. elegans. Current Biology, 2012,22(20):1938-1943.
pmid: 22981770 |
| [12] | BUCHER G, SCHOLTEN J, KLINGLER M. Parental RNAi in Tribolium (Coleoptera). Current Biology, 2002,12(3):R85-R86. |
| [13] |
XU H J, CHEN T, MA X F, XUE J, PAN P L, ZHANG X C, CHENG J A, ZHANG C X. Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Molecular Biology, 2013,22(6):635-647.
doi: 10.1111/imb.12051 pmid: 23937246 |
| [14] | CHENG L, SHARPLES R A, SCICLUNA B J, HILL A F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. Journal of Extracellular Vesicles, 2014,3:23743. |
| [15] |
VAN DER POL E, BOING A N, HARRISON P, STURK A, NIEUWLAND R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacological Reviews, 2012,64(3):676-705.
doi: 10.1124/pr.112.005983 pmid: 22722893 |
| [16] | ZHANG T, ZHAO Y L, ZHAO J H, WANG S, JIN Y, CHEN Z Q, FANG Y Y, HUA C L, DING S W, GUO H S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nature Plants, 2016,2(10):16153. |
| [17] |
SANNIGRAHI M K, SHARMA R, SINGH V, PANDA N K, RATTAN V, KHULLAR M. Role of host miRNA Hsa-miR-139-3p in HPV-16-induced carcinomas. Clinical Cancer Research, 2017,23(14):3884-3895.
doi: 10.1158/1078-0432.CCR-16-2936 pmid: 28143871 |
| [18] | HUANG Q, LI W, CHEN Y, RETSCHNIG-TANNE G, YANEZ O, NEUMANN P, EVANS J D. Dicer regulates Nosema ceranae proliferation in honeybees. Insect Molecular Biology, 2019,28(1):74-85. |
| [19] | 付中民, 陈华枝, 刘思亚, 祝智威, 范小雪, 范元婵, 万洁琦, 张璐, 熊翠玲, 徐国钧, 陈大福, 郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答. 中国农业科学, 2019,52(17):3069-3082. |
| FU Z M, CHEN H Z, LIU S Y, ZHU Z W, FAN X X, FAN Y C, WAN J Q, ZHANG L, XIONG C L, XU G J, CHEN D F, GUO R. Immune responses of Apis mellifera ligustia to Nosema ceranae stress. Scientia Agricultura Sinica, 2019,52(17):3069-3082. (in Chinese) | |
| [20] | 熊翠玲, 陈华枝, 祝智威, 王杰, 范小雪, 蒋海宾, 范元婵, 万洁琦, 卢家轩, 郑燕珍, 付中民, 徐国钧, 陈大福, 郭睿. 基于small RNA组学分析揭示意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答机制. 微生物学报, 2020,60(7):1458-1478. |
| XIONG C L, CHEN H Z, ZHU Z W, WANG J, FAN X X, JIANG H B, FAN Y C, WAN J Q, LU J X, ZHENG Y Z, FU Z M, XU G J, CHEN D F, GUO R. Unraveling the mechanism underlying the immune responses of Apis mellifera ligustica to Nosema ceranae stress based on small RNA omics analyses. Acta Microbiologica Sinica, 2020,60(7):1458-1478. (in Chinese) | |
| [21] | CHEN D F, CHEN H Z, DU Y, ZHOU D D, GENG S H, WANG H P, WAN J Q, XIONG C L, ZHENG Y Z, GUO R. Genome-wide identification of long non-coding RNAs and their regulatory networks involved in Apis mellifera ligustica response to Nosema ceranae infection. Insects, 2019,10(8):245. |
| [22] | 熊翠玲, 耿四海, 周丁丁, 石彩云, 郭意龙, 陈大福, 郑燕珍, 徐国钧, 张曦, 郭睿. 感染意大利蜜蜂工蜂的东方蜜蜂微孢子虫及其纯化孢子的高表达基因分析. 上海交通大学学报(农业科学版), 2019,37(2):6-13. |
| XIONG C L, GENG S H, ZHOU D D, SHI C Y, GUO Y L, CHEN D F, ZHENG Y Z, XU G J, ZHANG X, GUO R. Analysis of highly expressed genes in Nosema ceranae infecting the midguts of Apis mellifera ligustica worker and purified fungal spores. Journal of Shanghai Jiaotong University (Agricultural Science), 2019,37(2):6-13. (in Chinese) | |
| [23] | 周倪红, 王海朋, 周丁丁, 付中民, 祝智威, 范元婵, 张曦, 熊翠玲, 郑燕珍, 陈大福, 郭睿. 意大利蜜蜂工蜂中肠响应东方蜜蜂微孢子虫胁迫的可变剪接基因分析. 福建农林大学学报(自然科学版), 2020,49(3):372-379. |
| ZHOU N H, WANG H P, ZHOU D D, FU Z M, ZHU Z W, FAN Y C, ZHANG X, XIONG C L, ZHENG Y Z, CHEN D F, GUO R. Analysis on the response of alternatively splicing genes in Apis mellifera ligustica workers’ midguts to Nosema ceranae stress. Journal of Fujian Agriculture and Forestry University (Natural Science Edition), 2020,49(3):372-379. (in Chinese) | |
| [24] | 耿四海, 周丁丁, 范小雪, 蒋海宾, 祝智威, 王杰, 范元婵, 王心蕊, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 转录组分析揭示东方蜜蜂微孢子虫侵染意大利蜜蜂的分子机制. 昆虫学报, 2020,63(3):294-308. |
| GENG S H, ZHOU D D, FAN X X, JIANG H B, ZHU Z W, WANG J, FAN Y C, WANG X R, XIONG C L, ZHENG Y Z, FU Z M, CHEN D F, GUO R. Transcriptomic analysis reveals the molecular mechanism underlying Nosema ceranae infection of Apis mellifera ligustica. Acta Entomologica Sinica, 2020,63(3):294-308. (in Chinese) | |
| [25] | 耿四海, 石彩云, 范小雪, 王杰, 祝智威, 蒋海宾, 范元婵, 陈华枝, 杜宇, 王心蕊, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 微小RNA介导东方蜜蜂微孢子虫侵染意大利蜜蜂工蜂的分子机制. 中国农业科学, 2020,53(15):3187-3204. |
| GENG S H, SHI C Y, FAN X X, WANG J, ZHU Z W, JIANG H B, FAN Y C, CHEN H Z, DU Y, WANG X R, XIONG C L, ZHENG Y Z, FU Z M, CHEN D F, GUO R. The mechanism underlying microRNAs-mediated Nosema ceranae infection to Apis mellifera ligustica worker. Scientia Agricultura Sinica, 2020,53(15):3187-3204. (in Chinese) | |
| [26] | 郭睿, 杜宇, 熊翠玲, 郑燕珍, 付中民, 徐国钧, 王海朋, 陈华枝, 耿四海, 周丁丁, 石彩云, 赵红霞, 陈大福. 意大利蜜蜂幼虫肠道发育过程中的差异表达microRNA及其调控网络. 中国农业科学, 2018,51(21):4197-4209. |
| GUO R, DU Y, XIONG C L, ZHENG Y Z, FU Z M, XU G J, WANG H P, CHEN H Z, GENG S H, ZHOU D D, SHI C Y, ZHAO H X, CHEN D F. Differentially expressed microRNA and their regulation networks during the developmental process of Apis mellifera ligustica larval gut. Scientia Agricultura Sinica, 2018,51(21):4197-4209. (in Chinese) | |
| [27] | LANGMEAD B, TRAPNELL C, POP M, SALZBERG S L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 2009,10(3):R25. |
| [28] |
FRIEDLANDER M R, MACKOWIAK S D, LI N, CHEN W, RAJEWSKY N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research, 2012,40(1):37-52.
doi: 10.1093/nar/gkr688 pmid: 21911355 |
| [29] |
CHEN H Z, DU Y, XIONG C L, ZHENG Y Z, CHEN D F, GUO R. A comprehensive transcriptome data of normal and Nosema ceranae- stressed midguts of Apis mellifera ligustica workers. Data in Brief, 2019,26:104349.
pmid: 31516938 |
| [30] | GUO R, CHEN D F, XIONG C L, HOU C S, ZHENG Y Z, FU Z M, LIANG Q, DIAO Q Y, ZHANG L, WANG H Q, HOU Z X, KUMAR D. First identification of long non-coding RNAs in fungal parasite Nosema ceranae. Apidologie, 2018,49:660-670. |
| [31] |
ROBINSON M D, MCCARTHY D J, SMYTH G K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010,26(1):139-140.
doi: 10.1093/bioinformatics/btp616 pmid: 19910308 |
| [32] |
ALLEN E, XIE Z X, GUSTAFSON A M, CARRINGTON J C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 2005, 121(2): 207-221.
pmid: 33857425 |
| [33] |
SMOOT M E, ONO K, RUSCHEINSKI J, WANG P L, IDEKER T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics, 2011, 27(3): 431-432.
pmid: 33876181 |
| [34] | HUANG Q, CHEN Y P, WANG R W, CHENG S, EVANS J D. Host-parasite interactions and purifying selection in a microsporidian parasite of honey bees. PLoS ONE, 2016,11(2):e0147549. |
| [35] |
CORNMAN R S, CHEN Y P, SCHATZ M C, STREET C, ZHAO Y, DESANY B, EGHOLM M, HUTCHISON S, PETTIS J S, LIPKIN W I, EVANS J D. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathogens, 2009,5(6):e1000466.
pmid: 19503607 |
| [36] |
PALDI N, GLICK E, OLIVA M, ZILBERBERG Y, AUBIN L, PETTIS J, CHEN Y P, EVANS J D. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Applied and Environmental Microbiology, 2010,76(17):5960-5964.
pmid: 20622131 |
| [37] |
PELIN A, SELMAN M, ARIS-BROSOU S, FARINELLI L, CORRADI N. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environmental Microbiology, 2015,17(11):4443-4458.
doi: 10.1111/1462-2920.12883 pmid: 25914091 |
| [38] |
RODRÍGUEZ-GARCÍA C, EVANS J D, LI W, BRANCHICCELA B, LI J H, HEERMAN M C, BANMEKE O, ZHAO Y, HAMILTON M, HIGES M, MARTÍN-HERNÁNDEZ R, CHEN Y P. Nosemosis control in European honey bees, Apis mellifera, by silencing the gene encoding Nosema ceranae polar tube protein 3. Journal of Experimental Biology, 2018, 221(19): jeb184606.
pmid: 16363121 |
| [39] |
LIU H, LI M, HE X, CAI S, HE X, LU X. Transcriptome sequencing and characterization of ungerminated and germinated spores of Nosema bombycis. Acta Biochimica et Biophysica Sinica, 2016,48(3):246-256.
doi: 10.1093/abbs/gmv140 pmid: 26837419 |
| [40] | CAI Y, SHEN J. Modulation of host immune responses to Toxoplasma gondii by microRNAs. Parasite Immunology, 2017,39(2):12417. |
| [41] | ENTWISTLE L J, WILSON M S. MicroRNA-mediated regulation of immune responses to intestinal helminth infections. Parasite Immunology, 2017,39(2):e12406. |
| [42] |
GARBIAN Y, MAORI E, KALEV H, SHAFIR S, SELA I. Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathogens, 2012,8(12):e1003035.
pmid: 23308063 |
| [43] |
VIDAU C, PANEK J, TEXIER C, BIRON D G, BELZUNCES L P, GALL M L, BROUSSARD C, DELBAC F, ALAOUI H E. Differential proteomic analysis of midguts from Nosema ceranae-infected honeybees reveals manipulation of key host functions. Journal of Invertebrate Pathology, 2014,121:89-96.
doi: 10.1016/j.jip.2014.07.002 pmid: 25038465 |
| [44] | FIRE A, XU S, MONTGOMERY M K, KOSTAS S A, DRIVER S E, MELLO C C. Potent and specific genetic interference by double- stranded RNA in Caenorhabditis elegans. Nature, 1998, 391(6669): 806-811. |
| [45] |
HANNON G J. RNA interference. Nature, 2002,418(6894):244-251.
pmid: 12110901 |
| [46] | NDIKUMANA S, PELIN A, WILLIOT A, SANDERS J L, KENT M, CORRADI N. Genome analysis of Pseudoloma neurophilia: A microsporidian parasite of Zebrafish (Danio rerio). Journal of Eukaryotic Microbiology, 2017,64(1):18-30. |
| [47] | 鲁兴萌, 汪方炜. 家蚕肠球菌对微孢子虫体外发芽的抑制作用. 蚕业科学, 2002,28(2):126-128. |
| LU X M, WANG F W. Inhibition of cultured supernatant of enterococci strains on germination of Nosema bombycis spores in vitro. Acta Sericologica Sinica, 2002,28(2):126-128. (in Chinese) | |
| [48] |
YANG D L, PAN L X, PENG P, DANG X Q, LI C F, LI T, LONG M X, CHEN J, WU Y J, DU H H, et al. Interaction between SWP9 and polar tube proteins of the microsporidian Nosema bombycis and function of SWP9 as a scaffolding protein contribute to polar tube tethering to the spore wall. Infection and Immunity, 2017,85(3):e00872-16.
pmid: 28031263 |
| [49] |
WEIS W, BROWN J H, CUSACK S, PAULSON J C, SKEHEL J J, WILEY D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature, 1988,333(6172):426-431.
doi: 10.1038/333426a0 pmid: 3374584 |
| [50] | RUOSLAHTI E, PIERSCHBACHER M D. New perspectives in cell adhesion: RGD and integrins. Science, 1987,238(4826):491-497. |
| [51] |
LIU H, LI M, CAI S, HE X, SHAO Y, LU X. Ricin-B-lectin enhances microsporidia Nosema bombycis infection in BmN cells from silkworm Bombyx mori. Acta Biochimica et Biophysica Sinica, 2016,48(11):1050-1057.
doi: 10.1093/abbs/gmw093 pmid: 27649890 |
| [52] |
刘天明, 申玉龙, 刘庆军, 刘波. 古菌独特的脱氧酮糖酸(ED)葡萄糖酵解途径. 微生物学报, 2008,48(8):1126-1131.
pmid: 18956766 |
|
LIU T M, SHEN Y L, LIU Q J, LIU B. The unique Entner-Doudoroff (ED) glycolysis pathway of glucose in Archaea—A review. Acta Microbiologica Sinica, 2008,48(8):1126-1131. (in Chinese)
pmid: 18956766 |
|
| [53] |
HIGGINS C F. ABC transporters: From microorganisms to man. Annual Review of Cell Biology, 1992,8:67-113.
pmid: 1282354 |
| [54] |
HAMEL L P, NICOLE M C, DUPLESSIS S, ELLIS B E. Mitogen- activated protein kinase signaling in plant-interacting fungi: Distinct messages from conserved messengers. The Plant Cell, 2012,24(4):1327-1351.
pmid: 22517321 |
| [55] | 郭睿, 陈大福, 黄枳腱, 梁勤, 熊翠玲, 徐细建, 郑燕珍, 张曌楠, 解彦玲, 童新宇, 侯志贤, 江亮亮, 刀晨. 球囊菌胁迫中华蜜蜂幼虫肠道过程中病原的转录组学研究. 微生物学报, 2017,57(12):1865-1878. |
| GUO R, CHEN D F, HUANG Z J, LIANG Q, XIONG C L, XU X J, ZHENG Y Z, ZHANG Z N, XIE Y L, TONG X Y, HOU Z X, JIANG L L, DAO C. Transcriptome analysis of Ascosphaera apis stressing larval gut of Apis cerana cerana. Acta Microbiologica Sinica, 2017,57(12):1865-1878. (in Chinese) | |
| [56] | 陈大福, 郭睿, 熊翠玲, 梁勤, 郑燕珍, 徐细建, 黄枳腱, 张曌楠, 张璐, 李汶东, 童新宇, 席伟军. 胁迫意大利蜜蜂幼虫肠道的球囊菌的转录组分析. 昆虫学报, 2017,60(4):401-411. |
| CHEN D F, GUO R, XIONG C L, LIANG Q, ZHENG Y Z, XU X J, HUANG Z J, ZHANG Z N, ZHANG L, LI W D, TONG X Y, XI W J. Transcriptomic analysis of Ascosphaera apis stressing larval gut of Apis mellifera ligustica (Hyemenoptera: Apidae). Acta Entomologica Sinica, 2017,60(4):401-411. (in Chinese) |
| [1] | 王荣华,孟丽峰,冯毛,房宇,魏俏红,马贝贝,钟未来,李建科. 蜂王浆高产蜜蜂与意大利蜜蜂哺育蜂唾液腺蛋白质组分析[J]. 中国农业科学, 2022, 55(13): 2667-2684. |
| [2] | 冯睿蓉,付中民,杜宇,张文德,范小雪,王海朋,万洁琦,周紫彧,康育欣,陈大福,郭睿,史培颖. 中华蜜蜂幼虫肠道中微小RNA的鉴定及分析[J]. 中国农业科学, 2022, 55(1): 208-218. |
| [3] | 陈华枝,范元婵,蒋海宾,王杰,范小雪,祝智威,隆琦,蔡宗兵,郑燕珍,付中民,徐国钧,陈大福,郭睿. 基于纳米孔全长转录组数据完善东方蜜蜂微孢子虫的基因组注释[J]. 中国农业科学, 2021, 54(6): 1288-1300. |
| [4] | 王雍,李思妍,何思锐,张迪,连帅,王建发,武瑞. BLV-miRNA跨界调控人类靶基因预测及生物信息学分析[J]. 中国农业科学, 2021, 54(3): 662-674. |
| [5] | 高艳,朱雅楠,李秋方,苏松坤,聂红毅. 转录组学分析意大利蜜蜂脑部哺育行为相关基因[J]. 中国农业科学, 2020, 53(19): 4092-4102. |
| [6] | 陈华枝,祝智威,蒋海宾,王杰,范元婵,范小雪,万洁琦,卢家轩,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中微小RNA及其靶mRNA的比较分析[J]. 中国农业科学, 2020, 53(17): 3606-3619. |
| [7] | 耿四海,石彩云,范小雪,王杰,祝智威,蒋海宾,范元婵,陈华枝,杜宇,王心蕊,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 微小RNA介导东方蜜蜂微孢子虫侵染意大利蜜蜂工蜂的分子机制[J]. 中国农业科学, 2020, 53(15): 3187-3204. |
| [8] | 杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿. 微小RNA及其介导的竞争性内源RNA调控网络在意大利蜜蜂工蜂中肠发育过程中的潜在作用[J]. 中国农业科学, 2020, 53(12): 2512-2526. |
| [9] | 周丁丁,史小玉,王杰,范元婵,祝智威,蒋海宾,范小雪,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 东方蜜蜂微孢子虫孢子中长链非编码RNA的竞争性内源RNA调控网络及潜在功能[J]. 中国农业科学, 2020, 53(10): 2122-2136. |
| [10] | 杜宇,周丁丁,万洁琦,卢家轩,范小雪,范元婵,陈恒,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 意大利蜜蜂工蜂中肠发育过程中的差异基因 表达谱及调控网络[J]. 中国农业科学, 2020, 53(1): 201-212. |
| [11] | 薛亚东,杨露,杨慧丽,李冰,林亚楠,张怀胜,郭战勇,汤继华. 玉米C型细胞质雄性不育花药不同发育时期的转录组分析[J]. 中国农业科学, 2019, 52(8): 1308-1323. |
| [12] | 付中民,陈华枝,刘思亚,祝智威,范小雪,范元婵,万洁琦,张璐,熊翠玲,徐国钧,陈大福,郭睿. 意大利蜜蜂响应东方蜜蜂微孢子虫胁迫的免疫应答[J]. 中国农业科学, 2019, 52(17): 3069-3082. |
| [13] | 于静,张卫星,马兰婷,胥保华. 饲粮α-亚麻酸水平对意大利蜜蜂工蜂幼虫生理机能的影响[J]. 中国农业科学, 2019, 52(13): 2368-2378. |
| [14] | 郭睿,杜宇,童新宇,熊翠玲,郑燕珍,徐国钧,王海朋,耿四海,周丁丁,郭意龙,吴素珍,陈大福. 意大利蜜蜂幼虫肠道在球囊菌侵染前期的 差异表达microRNA及其调控网络[J]. 中国农业科学, 2019, 52(1): 166-180. |
| [15] | 郭睿,陈华枝,熊翠玲,郑燕珍,付中民,徐国钧,杜宇,王海朋,耿四海,周丁丁,刘思亚,陈大福. 意大利蜜蜂工蜂中肠发育过程中的差异表达环状RNA及其调控网络分析[J]. 中国农业科学, 2018, 51(23): 4575-4590. |
|
||