













中国农业科学 ›› 2020, Vol. 53 ›› Issue (7): 1381-1396.doi: 10.3864/j.issn.0578-1752.2020.07.008
贾海燕1,宋丽云1,徐翔1,解屹1,张超群2,刘天波3,赵存孝4,申莉莉1,王杰1,李莹1,王凤龙1,杨金广1
收稿日期:2019-09-16
接受日期:2019-11-06
出版日期:2020-04-01
发布日期:2020-04-14
作者简介:贾海燕,E-mail:基金资助:HaiYan JIA1,LiYun SONG1,Xiang XU1,Yi XIE1,ChaoQun ZHANG2,TianBo LIU3,CunXiao ZHAO4,LiLi SHEN1,Jie WANG1,Ying LI1,FengLong WANG1,JinGuang YANG1
Received:2019-09-16
Accepted:2019-11-06
Online:2020-04-01
Published:2020-04-14
摘要: 【目的】 筛选不同温度下烟草花叶病毒(Tobacco mosaic virus,TMV)侵染后枯斑三生烟(Nicotiana tabacum var. Samsun NN)差异表达的长链非编码RNA(long non-coding RNA,lncRNA),研究lncRNA在枯斑三生烟抗性反应中的作用。【方法】 N基因的温度敏感性使枯斑三生烟在25℃时具备对TMV的抗性、在31℃抗性丧失,在这两个温度条件下对枯斑三生烟接种TMV和磷酸盐缓冲盐水(phosphate buffered saline,PBS),48 h后提取系统叶总RNA,构建链特异性文库后进行深度测序。对测序结果进行过滤后利用HTSeq将有效数据与近缘品种TN90(N. tabacum var. TN90)基因组比对,筛选得到lncRNA后利用FPKM法估计lncRNA的表达水平。通过edgeR筛选差异表达lncRNA(differentially expressed lncRNA,DElncRNA),并利用qRT-PCR技术对这一结果进行验证。通过共定位及共表达分析预测DElncRNA的靶基因,通过参考基因组注释、GO和KEGG富集分析研究靶基因的功能。【结果】 4个处理共12个样本经lncRNA-seq各测得约8 000万条clean reads,共获得4 737条已知lncRNA、40 169条新lncRNA。其中64个lncRNA在不同温度条件下TMV侵染后存在差异表达,qRT-PCR测定结果显示这些lncRNA的测序正确率在80%左右,表明本研究所得测序数据具备较高的可信度。对DElncRNA进行靶基因预测,发现一些基因同时被25℃下调和31℃上调的DElncRNA靶向。靶基因注释功能丰富,主要参与植物抗病、激素和代谢等生理过程。部分可能与激素通路相关的lncRNA,在25℃下TMV侵染时呈现下调趋势,而在31℃下TMV侵染则呈现上调趋势。GO富集分析显示靶基因主要参与构成膜、囊泡等组分,具备钙、钾离子通道抑制剂活性等分子功能,使相应离子得以转运引发随后的反应,同时也参与发病、抗原加工和呈现、细胞分裂素代谢等生理过程。KEGG分析发现靶基因显著富集在植物激素信号转导通路,25℃下调和31℃上调的DElncRNA靶基因同时富集在激素信号传导、ABC运输蛋白、苯丙烷类生物合成等通路。【结论】 不同温度(25℃和31℃)条件下TMV侵染枯斑三生烟后,长链非编码RNA差异表达,DElncRNA通过作用于激素信号传导、物质转运等过程参与寄主系统获得性抗性反应。研究结果可为揭示植物系统获得性抗性中lncRNA的调控功能以及新型抗病毒技术开发提供依据。
贾海燕,宋丽云,徐翔,解屹,张超群,刘天波,赵存孝,申莉莉,王杰,李莹,王凤龙,杨金广. 不同温度下TMV侵染枯斑三生烟的LncRNA差异表达[J]. 中国农业科学, 2020, 53(7): 1381-1396.
HaiYan JIA,LiYun SONG,Xiang XU,Yi XIE,ChaoQun ZHANG,TianBo LIU,CunXiao ZHAO,LiLi SHEN,Jie WANG,Ying LI,FengLong WANG,JinGuang YANG. Differential Expression of LncRNAs in Nicotiana tabacum var. Samsun NN Infected by TMV at Different Temperatures[J]. Scientia Agricultura Sinica, 2020, 53(7): 1381-1396.

图1
不同温度下TMV接种前后的枯斑三生烟 A:25℃条件下TMV接种前的枯斑三生烟Samsun NN before TMV inoculation at 25℃;B:25℃条件下TMV接种48 h后的枯斑三生烟Samsun NN after 48 hours of TMV inoculation at 25℃;C:31℃条件下TMV接种前的枯斑三生烟Samsun NN before inoculation of TMV at 31℃;D:31℃条件下TMV接种48 h后的枯斑三生烟Samsun NN after 48 hours of TMV inoculation at 31℃。标注为小写字母的图片展示相应枯斑三生烟的接种叶The pictures marked in lowercase letters show the inoculated leaves of the corresponding tobacco"

表1
25℃下差异表达lncRNA引物序列"
| 转录本ID Transcript ID | 基因ID Gene ID | 正向引物序列 Forward primer sequence (5′ to 3′) | 反向引物序列 Reverse primer sequence (5′ to 3′) |
|---|---|---|---|
| LNC_000098 | XLOC_000472 | CCCTCCACGCAGTTCTTCTGA | TGCCGGGAGTTTCGGGTTTA |
| LNC_000799 | XLOC_003050 | GCCTTCTGCCGAATTGTTGG | GCTCAAAGGAGGCCAAGTCC |
| LNC_001093 | XLOC_004138 | ATGCGTCAGTTTGCATGGGA | GAAATCTCAAGGGTAGCTGTACCA |
| LNC_006130 | XLOC_023441 | GCCTTCGACTTGTCGTTTGCT | ATCCGCAAATCAGCCCTTCC |
| LNC_012986 | XLOC_049416 | CCTCTGCCTCAGGCTGTCTCG | TTCTGCCGCTTCCGTTATTGCTG |
| LNC_019285 | XLOC_073034 | GGATTGAATCTAAGATGAATGCTTGGT | ACAGTCACAAGGGTCTAGAAGG |
| LNC_028742 | XLOC_108482 | ACGCTGGCGAACGATAATAGAACC | ACGGTCGAGATTCTCCTGGTCAG |
| LNC_037710 | XLOC_141968 | TCTCTTTCTACCGGGAATTTAAAAAGT | CCATTACGTATTGATTGGTGAGCT |
| XR_001643474.1 | 107765638 | CCACTTGCATGGGCAGTGAT | GCCACCACACCAACTTCCTC |
| XR_001643621.1 | 107766213 | CGGAGAGGTGGAATGGCAAGTG | TCCGAGATGTGCCTCCAAGACC |
| XR_001644424.1 | 107769516 | AACATTCACGGCCAGCAGAACC | CTGAAGGCGCGACTGAGATTAGC |
| XR_001645141.1 | 107772701 | TCATTCTCAGGTCCTCCGTTGT | TCTCTCTGTGCTTTCCGCCT |
| XR_001650509.1 | 107796801 | CGCCCATGTACCCGATTCTG | CTTGTTCCAGATGCGCCAGT |
| XR_001654053.1 | 107812073 | CACTGCCTACACAACCTACACGTC | AACAACATGCTGCGAAGAGACTCC |
| XR_001657220.1 | 107826167 | AATGGCTTTGTCTGCCTCGG | AGGCGCGTTGATGAAAGAGG |
| LNC_001159 | XLOC_004417 | GACACAGGTACTTGGTTCGCTC | AAGAACCGGCACAATACGCC |
| LNC_003116 | XLOC_011996 | GGTTTGCAGCGATAACTCGG | TGGCTTCAACTATTCCCGCA |
| LNC_004594 | XLOC_017602 | AGTGAATGTAACGGAGGCAGCAAC | ACACAGAAGAGGATCGAGGTCGTC |
| LNC_015076 | XLOC_057288 | CTCTTGCAGGAAGGTTGGCT | CCTCATACGGCTCCTCCCTT |
| LNC_015077 | XLOC_057288 | CTCTTGCAGGAAGGTTGGCT | CCTCATACGGCTCCTCCCTT |
| LNC_028743 | XLOC_108482 | ACGCTGGCGAACGATAATAGAACC | ACGGTCGAGATTCTCCTGGTCAG |
| LNC_030105 | XLOC_113591 | TCCCAACGTGTTACATAAGGCA | GCACGGTGTTCTTCGACTCA |
| LNC_036321 | XLOC_136711 | CTCTGTCATTCTTCGGTGCCGATG | TGAGCTGCCATGCCATGATACAAG |
| LNC_038081 | XLOC_143307 | GCTTAGTGTGTGACTCGTTGGT | TGCGAAGTGATGGGTTGGTT |
| XR_001643840.1 | 107767123 | GTTAGTTCGAGGCTGCGGTCAAG | CATCGGCGGCGGACAATTACC |
| XR_001646643.1 | 107779549 | GGTTGGATCAGCAGCAGCAGTAG | AGCAGCAGTCTCATTAGCAGCAAC |
| XR_001648910.1 | 107789767 | ATCCGAACGACCACTCCCAG | TTGCAAGTCATCACCGTCCG |
| XR_001649452.1 | 107792124 | TGTTGGAGAAGGCTGGGACT | CGGCATCTGCTGCTTCTAGT |
| XR_001651797.1 | 107802252 | GGCATCAATCACACATGCCGT | AGCAGCCACTGACTTGGAAAC |
| XR_001654265.1 | 107813134 | GCACTCGAACCAGAAGGCAA | CAGCAGAAGCTCGACCACAA |
| XR_001654587.1 | 107814577 | GTTCATCGCCTTCTTCTGCCTCTC | AAGCACCGAGAGCAACCAACATAG |
| XR_001655076.1 | 107816953 | AATGGCTTTGTCTGCCTCGG | AGGCGCGTTGATGAAAGAGG |
| XR_001657217.1 | 107826167 | GTGGATGGCGAATGCGATCT | CTGAGGTCCAGCTGCCAATG |
| XR_001657841.1 | 107829108 | GTGGTGGTGCATGGCCGTTC | TAGCAGGCTGAGGTCTCGTTCG |
| XR_001658459.1 | 107831913 | GTGGTGGTGCATGGCCGTTC | TAGCAGGCTGAGGTCTCGTTCG |
表2
31℃下差异表达lncRNA引物序列"
| 转录本ID Transcript ID | 基因ID Gene ID | 正向引物序列 Forward primer sequence (5′ to 3′) | 反向引物序列 Reverse primer sequence (5′ to 3′) |
|---|---|---|---|
| LNC_008396 | XLOC_031876 | GCGATCTGCCGAAGCTGTGG | TGCGTTACTCAAGCCGACATTCTC |
| LNC_008562 | XLOC_032544 | TCCTGTTGCTTCATTGCTGCT | ACAGATGAACACAGCGCAGG |
| LNC_008815 | XLOC_033553 | GACTGTGAAACTGCGAATGGC | GCATCCCTTCCAGAAGTCGG |
| LNC_010207 | XLOC_038635 | AGACGAACAACTGCGAAAGCA | CTGGTCGGCATCGTTTATGGT |
| LNC_013541 | XLOC_051547 | AGCACCATCCTCACTTCCACCAG | CGCAGGCTGCCTCACAACTTG |
| LNC_016042 | XLOC_060869 | GATTAAGACAGCAGGACGGTGGTC | GGCTAGTTGATTCGGCAGGTGAG |
| LNC_018096 | XLOC_068634 | AGCCAAGCGTTCATAGCGAC | AACCCAGCTCACGTTCCCTA |
| LNC_024626 | XLOC_093025 | ACCACCTGTGGCTCCAGTTACC | TAGGCGGACCTCGCAGAATCTTAG |
| LNC_033085 | XLOC_124537 | AGCGAGCAGTCTGGACTCCTATG | ACGAGTAACCAGCGCAATTGGAG |
| LNC_033563 | XLOC_126355 | GTGGTGGTGCATGGCCGTTC | TAGCAGGCTGAGGTCTCGTTCG |
| LNC_036321 | XLOC_136711 | CTCTGTCATTCTTCGGTGCCGATG | TGAGCTGCCATGCCATGATACAAG |
| LNC_037998 | XLOC_143050 | GATTAAGACAGCAGGACGGTGGTC | GGCTAGTTGATTCGGCAGGTGAG |
| LNC_038712 | XLOC_145744 | CAACCGCTCAGCCATCTCTC | GGGGTATCCACCCCTATGGC |
| XR_001648128.1 | 107786374 | GGAGGGAAGCTGGGTCGTAT | GGCAATTCTCCATCGGCTCC |
| XR_001655221.1 | 107817578 | GAGAGTCGGGCTGCAACTTC | GGGAAGCATGGCACCAACAA |
| XR_001655293.1 | 107817783 | GTCTGAGGTTGCGGTGAAGGC | ATTGGCGGCAGAGAATTGACAGAG |
| XR_001656793.1 | 107824145 | TCTCATGTGGGATGGCACGA | CATTCCACTGCGCACCTCTC |
| XR_001657266.1 | 107826393 | TTGAGGCCACTGTTCAAGACTTGG | CGCGAGCAAGGTCATCAGAGC |
| LNC_018541 | XLOC_070302 | ACTTCACAAGCAGCAGCTAGTTCC | GTAATGCGCCAGGTGCCGTAG |
| LNC_034892 | XLOC_131403 | GTGGGTTCCAGACCACAACC | AGATGGAAGGGGCAGGTGTT |
| XR_001644489.1 | 107769825 | CCGTGGACGTGACAACATTGGAG | CGGTGACTGTCGCTGAGATACTTG |
| XR_001646112.1 | 107776988 | TCGCTGCTGATTGTTGCTGTCTC | GACTCGTCGGCAACCTCAACTG |
| XR_001647877.1 | 107785495 | ATCAATGTGGGAGCTTGCCT | GCTTGTGCGTGTCTCGACTA |
| XR_001647999.1 | 107785872 | GCGGATGAGGTATGGTTCACAGC | AGCTGCTTCCATAGTCTGTTGCTG |
| XR_001648126.1 | 107786374 | ATGAGAGTGGAGTGGGGCAG | TGCGCCCAATAGGTTATGCG |
| XR_001649356.1 | 107791865 | ACGTTGCTTTACACTTTACCCTG | TGGAGGAAGAAACCAAGCGT |
| XR_001654070.1 | 107812109 | TCCGTTCAAGTCCACCTGAAGTTC | GCAGCTCACCGTGGAAGTCTC |
| XR_001656841.1 | 107824342 | CAAGCCATCATGCCTCACAGT | CCGGAGTCTACCACCCATTCT |
| XR_001656939.1 | 107824792 | CTGCTGGTCCAGAAGCCGTTAAG | AATGTCGTCACGAGTTCGGTCATG |
| XR_001658441.1 | 107831745 | CAGACGAGGGGTGGAGATGT | GCAAATGCCAACAAACAGGGA |
表3
测序数据概览"
| 样本名称 Sample name | 原始读段 Raw reads | 有效读段 Clean reads | 测序错误率 Error rate (%) | 可定位序列总数及占比 Total mapped | 多定位序列计数及占比 Multiple mapped | 单定位序列计数及占比 Uniquely mapped |
|---|---|---|---|---|---|---|
| PBS_25_1 | 81285872 | 80011202 | 0.02 | 76771937 (95.95%) | 36818251 (46.02%) | 39953686 (49.94%) |
| PBS_25_2 | 79744462 | 78438974 | 0.02 | 75316854 (96.02%) | 36751852 (46.85%) | 38565002 (49.17%) |
| PBS_25_3 | 82086686 | 81323058 | 0.02 | 78074493 (96.01%) | 38211722 (46.99%) | 39862771 (49.02%) |
| PBS_31_1 | 112775324 | 110071878 | 0.01 | 106555280 (96.81%) | 51925797 (47.17%) | 54629483 (49.63%) |
| PBS_31_2 | 110922928 | 108381110 | 0.01 | 105100267 (96.97%) | 51096062 (47.14%) | 54004205 (49.83%) |
| PBS_31_3 | 111456946 | 109144098 | 0.01 | 105971578 (97.09%) | 51922061 (47.57%) | 54049517 (49.52%) |
| TMV_25_1 | 82779880 | 81084374 | 0.02 | 77865206 (96.03%) | 38330300 (47.27%) | 39534906 (48.76%) |
| TMV_25_2 | 81397982 | 79800752 | 0.02 | 76525369 (95.90%) | 36994028 (46.36%) | 39531341 (49.54%) |
| TMV_25_3 | 84779126 | 83206166 | 0.02 | 79834756 (95.95%) | 40768694 (49.00%) | 39066062 (46.95%) |
| TMV_31_1 | 86454182 | 85145676 | 0.02 | 81734255 (95.99%) | 39993324 (46.97%) | 41740931 (49.02%) |
| TMV_31_2 | 82671238 | 81754880 | 0.01 | 78713311 (96.28%) | 35893618 (43.90%) | 42819693 (52.38%) |
| TMV_31_3 | 96258450 | 93993096 | 0.02 | 90712104 (96.51%) | 44629779 (47.48%) | 46082325 (49.03%) |
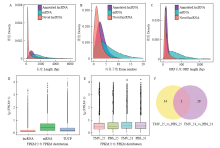
图2
LncRNA-seq结果分析 A:lncRNA和mRNA序列长度的密度分布Density distribution of sequence lengths of lncRNA and mRNA;B:lncRNA和mRNA外显子个数的密度分布Density distribution of exon number of lncRNA and mRNA;C:lncRNA和mRNA ORF长度的密度分布Density distribution of ORF length of lncRNA and mRNA;D:不同类型转录本表达水平比较Comparison of expression levels of different types of transcripts;E:不同处理转录本表达水平比较Comparison of expression levels of different treatments of transcripts;F:25℃和31℃比较组中差异表达lncRNA数目统计The numbers of differentially expressed lncRNAs in the comparison groups of 25℃ and 31℃"
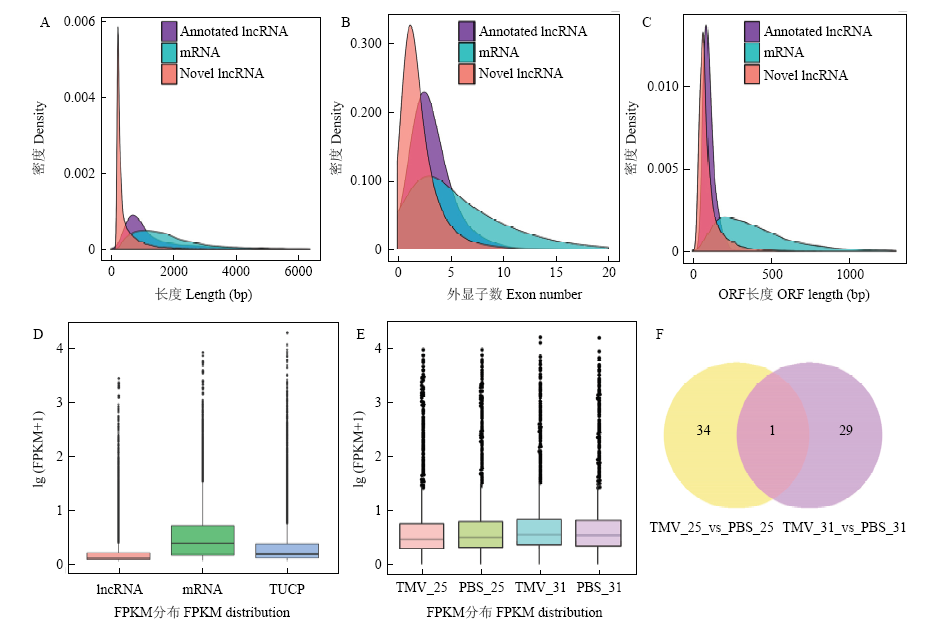
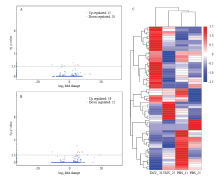
图3
差异表达lncRNA分析 A:25℃差异表达lncRNA火山图,横坐标代表转录本在不同样本中表达倍数变化,纵坐标代表转录本表达量变化差异的统计学显著性,q-value即P-adjust;图中红色点表示有显著性差异表达的上调转录本,蓝色点表示有显著性差异表达的下调转录本Volcano plot of differentially expressed lncRNAs at 25℃. The abscissa represents expression fold change in different samples, the ordinate represents the statistical significance of difference in transcript expression, and the q-value is P-adjust. The red dot in the figure indicates the up-regulated transcript with significant differential expression, and the blue dot indicates the down-regulated transcript with significant differential expression;B:31℃差异表达lncRNA火山图Volcano plot of differentially expressed lncRNAs at 31℃。C:差异表达lncRNA表达量层次聚类,以 lg (FPKM+1) 值进行聚类,红色代表基因高表达,蓝色表示低表达基因,颜色从红到蓝表示lg (FPKM+1)从大到小DElncRNA expression level hierarchical clustering, clustering was performed with lg (FPKM+1) values, with red representing high expression of genes and blue indicating low expressed genes, colors from red to blue indicate lg (FPKM+1) from large to small"
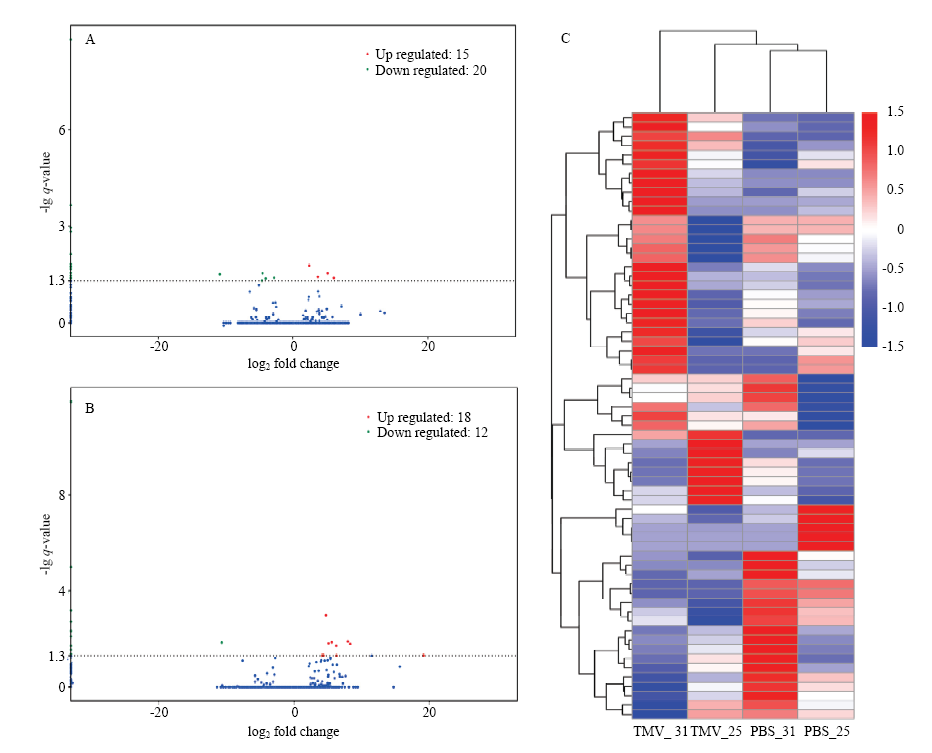
图4
25℃条件下差异表达lncRNA 的qRT-PCR验证 A:25℃和TMV处理下uDElncRNA的相对表达量Relative expression of up-regulated DElncRNAs after TMV treatment at 25℃;B:25℃和TMV处理下dDElncRNA的相对表达量Relative expression of down-regulated DElncRNAs after TMV treatment at 25℃ 误差线表示3组生物学重复的正负标准差,采用t-检验分析差异显著性 Error bars indicate the ±SD of three biological replicates. Student’s t-test. *:P<0.1; **:P<0.01; ***:P<0.001。图5同 The same as Fig. 5"
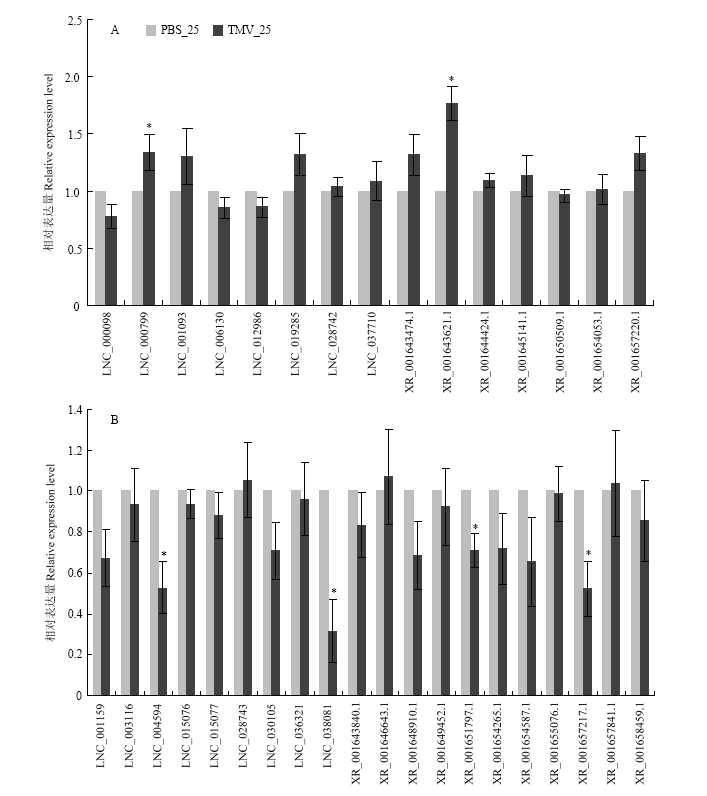
表4
GO条目富集的 DElncRNA靶基因数目"
| 条目类型 Term type | GO条目 GO term | 比较组合 Comparison group | 基因数 Number of genes |
|---|---|---|---|
| 生物过程 Biological process (BP) | 单组织进程Single-organism process | TMV_25_vs_PBS_25 | 133 |
| 单组织细胞进程Single-organism cellular process | TMV_25_vs_PBS_25 | 113 | |
| 氮化合物代谢过程Nitrogen compound metabolic process | TMV_25_vs_PBS_25 | 108 | |
| 有机环状化合物代谢过程Organic cyclic compound metabolic process | TMV_25_vs_PBS_25 | 105 | |
| 细胞氮化合物代谢过程Cellular nitrogen compound metabolic process | TMV_25_vs_PBS_25 | 104 | |
| 单组织进程Single-organism process | TMV_31_vs_PBS_31 | 21 | |
| 囊泡介导的运输Vesicle-mediated transport | TMV_31_vs_PBS_31 | 20 | |
| 细胞对刺激的反应Cellular response to stimulus | TMV_31_vs_PBS_31 | 9 | |
| 自RNA聚合酶II启动子转录Transcription from RNA polymerase II promoter | TMV_31_vs_PBS_31 | 8 | |
| 端粒维持Telomere maintenance | TMV_31_vs_PBS_31 | 7 | |
| 细胞组分 Cellular component (CC) | 细胞Cell | TMV_25_vs_PBS_25 | 188 |
| 细胞组分Cell part | TMV_25_vs_PBS_25 | 188 | |
| 细胞内组分Intracellular part | TMV_25_vs_PBS_25 | 166 | |
| 细胞内膜结合细胞器Intracellular membrane-bounded organelle | TMV_25_vs_PBS_25 | 114 | |
| 膜结合细胞器Membrane-bounded organelle | TMV_25_vs_PBS_25 | 114 | |
| 质膜组分Plasma membrane part | TMV_31_vs_PBS_31 | 8 | |
| 微管组织中心Microtubule organizing center | TMV_31_vs_PBS_31 | 4 | |
| 反式高尔基体转运囊泡膜Trans-Golgi network transport vesicle membrane | TMV_31_vs_PBS_31 | 2 | |
| 网格蛋白囊膜Clathrin vesicle coat | TMV_31_vs_PBS_31 | 2 | |
| 网格蛋白外壳Clathrin coat | TMV_31_vs_PBS_31 | 3 | |
| 分子功能 Molecular function (MF) | 阳离子结合Cation binding | TMV_25_vs_PBS_25 | 69 |
| 锌离子结合Zinc ion binding | TMV_25_vs_PBS_25 | 41 | |
| 糖基转移酶活性Transferase activity, transferring glycosyl groups | TMV_25_vs_PBS_25 | 31 | |
| 己糖基转移酶活性Transferase activity, transferring hexosyl groups | TMV_25_vs_PBS_25 | 23 | |
| DNA解旋酶活性DNA helicase activity | TMV_25_vs_PBS_25 | 13 | |
| UDP-糖基转移酶活性UDP-glycosyltransferase activity | TMV_31_vs_PBS_31 | 8 | |
| 抗氧化活性Antioxidant activity | TMV_31_vs_PBS_31 | 8 | |
| 过氧化物酶活性Peroxidase activity | TMV_31_vs_PBS_31 | 7 | |
| 作为受体作用于过氧化物的氧化还原酶活性 Oxidoreductase activity, acting on peroxide as acceptor | TMV_31_vs_PBS_31 | 7 | |
| 翻译监管活动Translation regulator activity | TMV_31_vs_PBS_31 | 5 |
表5
KEGG 通路富集的DElncRNA靶基因数目比较"
| 通路 Pathway | TMV_25_vs_PBS_25 | TMV_31_vs_PBS_31 |
|---|---|---|
| 植物激素信号转导Plant hormone signal transduction | 22 | 15 |
| 苯丙烷类生物合成Phenylpropanoid biosynthesis | 12 | 9 |
| mRNA监测途径mRNA surveillance pathway | 11 | 6 |
| 嘧啶代谢Pyrimidine metabolism | 10 | 6 |
| ABC运输蛋白ABC transporters | 3 | 3 |
| 非同源末端连接Non-homologous end-joining | 3 | 1 |
| RNA转运RNA transport | 6 | _ |
| 碱基切除修复Base excision repair | 5 | _ |
| 错配修复Mismatch repair | 4 | _ |
| 氨酰基-tRNA生物合成Aminoacyl-tRNA biosynthesis | 4 | _ |
| 缬氨酸、亮氨酸和异亮氨酸的生物合成Valine, leucine and isoleucine biosynthesis | 3 | _ |
| 缬氨酸、亮氨酸和异亮氨酸降解Valine, leucine and isoleucine degradation | 3 | _ |
| 同源重组Homologous recombination | 3 | _ |
| 一个叶酸碳库One carbon pool by folate | 3 | _ |
| 二萜生物合成Diterpenoid biosynthesis | 1 | _ |
| 苯丙氨酸代谢Phenylalanine metabolism | _ | 7 |
| 水泡运输中的SNARE相互作用SNARE interactions in vesicular transport | _ | 1 |
| 磷脂酰肌醇信号系统Phosphatidylinositol signaling system | _ | 1 |
| 肌醇磷酸代谢Inositol phosphate metabolism | _ | 1 |
| 酮体的合成和降解Synthesis and degradation of ketone bodies | _ | 1 |
| 丁酸代谢Butanoate metabolism | _ | 1 |
| 鞘脂代谢Sphingolipid metabolism | _ | 1 |
| [1] | WHITHAM S, DINESH-KUMAR S P, CHOI D, HEHL R, CORR C, BAKER B. The product of the Tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell, 1994,78(6):1101-1115. |
| [2] | WANG H V, CHEKANOVA J A . Long noncoding RNAs in plants//RAO M R S. Long Non Coding RNA Biology. Singapore: Springer, 2017: 133-154. |
| [3] | QUAN M, CHEN J, ZHANG D . Exploring the secrets of long noncoding RNAs. International Journal of Molecular Sciences, 2015,16(3):5467-5496. |
| [4] | RYALS J A, NEUENSCHWANDER U H, WILLITS M G, MOLINA A, STEINER H Y, HUNT M D . Systemic acquired resistance. The Plant Cell, 1996,8(10):1809-1819. |
| [5] | ERICKSON F L, HOLZBERG S, CALDERON‐URREA A, HANDLEY V, AXTELL M, CORR C, BAKER B. The helicase domain of the TMV replicase proteins induces the N‐mediated defence response in tobacco. The Plant Journal, 1999,18(1):67-75. |
| [6] | GAFFNEY T, FRIEDRICH L, VERNOOIJ B, NEGROTTO D, NYE G, UKNES S, WARD E, KESSMANN H, RYALS J . Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 1993,261(5122):754-756. |
| [7] | MUR L A, KENTON P, LLOYD A J, OUGHAM H, PRATS E . The hypersensitive response; the centenary is upon us but how much do we know? Journal of Experimental Botany, 2008,59(3):501-520. |
| [8] | YANG Y X, AHAMMED G J, WU C, FAN S Y, ZHOU Y H . Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Current Protein and Peptide Science, 2015,16(5):450-461. |
| [9] | BAKER B, ZAMBRYSKI P, STASKAWICZ B, DINESH-KUMAR S P. Signaling in plant-microbe interactions. Science, 1997,276(5313):726-733. |
| [10] | SMITH H B . Signal transduction in systemic acquired resistance. The Plant Cell, 2000,12(2):179-181. |
| [11] | CUTT J R, KLESSIG D F . Pathogenesis-Related Proteins: Genes Involved in Plant Defense. Vienna: Springer, 1992: 209-243. |
| [12] | 肖万福 . 烟草感染花叶病过程中差异表达基因与蛋白质的筛选及应用[D]. 郑州: 河南农业大学, 2017. |
| XIAO W F . Screening and application of differential expressed genes and proteins in tobacco mosaic disease[D]. Zhengzhou: Henan Agricultural University, 2017. (in Chinese) | |
| [13] | WANG J, YANG Y, JIN L, LING X, LIU T, CHEN T, JI Y, YU W, ZHANG B . Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biology, 2018,18:104. |
| [14] | YANG Y, LIU T, SHEN D, WANG J, LING X, HU Z, CHEN T, HU J, HUANG J, YU W, DOU D, WANG M B, ZHANG B . Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathogens, 2019,15(1):e1007534. |
| [15] | SEO J S, SUN H X, PARK B S, HUANG C H, YEH S D, JUNG C, CHUA N H . ELF18-induced long-noncoding RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. The Plant Cell, 2017,29(5):1024-1038. |
| [16] | SEO J S, DILOKNAWARIT P, PARK B S, CHUA N H . ELF18- induced long noncoding RNA 1 evicts fibrillarin from mediator subunit to enhance pathogenesis-related gene 1 (PR1) expression. New Phytologist, 2019,221(4):2067-2079. |
| [17] | ANDERS S . HTSeq: Analysing high-throughput sequencing data with Python[EB/OL]. . |
| [18] | PERTEA M, PERTEA G M, ANTONESCU C M, CHANG T C, MENDELL J T, SALZBERG S L . StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology, 2015,33(3):290-295. |
| [19] | TRAPNELL C, WILLIAMS B A, PERTEA G, MORTAZAVI A, KWAN G, VAN BAREN M J, SALZBERG S L, WOLD B J, PACHTER L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 2010,28(5):511-515. |
| [20] | SUN L, LUO H, BU D, ZHAO G, YU K, ZHANG C, LIU Y, CHEN R, ZHAO Y . Utilizing sequence intrinsic composition to classify protein- coding and long non-coding transcripts. Nucleic Acids Research, 2013,41(17):e166. |
| [21] | KANG Y J, YANG D C, KONG L, HOU M, MENG Y Q, WEI L, GAO G . CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Research, 2017,45(Web Server issue):W12-W16. |
| [22] | MISTRY J, BATEMAN A, FINN R D . Predicting active site residue annotations in the Pfam database. BMC Bioinformatics, 2007,8:298. |
| [23] | CABILI M N, TRAPNELL C, GOFF L, KOZIOL M, TAZON-VEGA B, REGEV A, RINN J L . Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes and Development, 2011,25(18):1915-1927. |
| [24] | ROBINSON M D, MCCARTHY D J, SMYTH G K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010,26(1):139-140. |
| [25] | LIVAK K J, SCHMITTGEN T D . Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method . Methods, 2001,25(4):402-408. |
| [26] | LI T, ZHANG G, WU P, DUAN L, LI G, LIU Q, WANG J . Dissection of myogenic differentiation signatures in chickens by RNA-Seq analysis. Genes, 2018,9(1):34. |
| [27] | WANG K C, CHANG H Y . Molecular mechanisms of long noncoding RNAs. Molecular Cell, 2011,43(6):904-914. |
| [28] | YOUNG M D, WAKEFIELD M J, SMYTH G K, OSHLACK A . Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biology, 2010,11(2):R14. |
| [29] | KANEHISA M, ARAKI M, GOTO S, HATTORI M, HIRAKAWA M, ITOH M, KATAYAMA T, KAWASHIMA S, OKUDA S, TOKIMATSU T, YAMANISHI Y . KEGG for linking genomes to life and the environment. Nucleic Acids Research, 2007,36(Database issue):D480-D484. |
| [30] | MAO X, CAI T, OLYARCHUK J G, WEI L . Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 2005,21(19):3787-3793. |
| [31] | OSATO N, SUZUKI Y, IKEO K, GOJOBORI T . Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics, 2007,176(2):1299-1306. |
| [32] | FAGHIHI M A, WAHLESTEDT C . Regulatory roles of natural antisense transcripts. Nature Reviews. Molecular Cell Biology, 2009,10(9):637-643. |
| [33] | BENJAMINS R, SCHERES B . Auxin: The looping star in plant development. Annual Review of Plant Biology, 2008,59:443-465. |
| [34] | WILSON M S, LIVERMORE T M, SAIARDI A . Inositol pyrophosphates: Between signalling and metabolism. Biochemical Journal, 2013,452(3):369-379. |
| [35] | SAHA M, SARKAR S, SARKAR B, SHARMA B K, BHATTACHARJEE S, TRIBEDI P . Microbial siderophores and their potential applications: A review. Environmental Science and Pollution Research, 2016,23(5):3984-3999. |
| [36] | ZHENG Y, DING B, FEI Z, WANG Y . Comprehensive transcriptome analyses reveal tomato plant responses to tobacco rattle virus-based gene silencing vectors. Scientific Reports, 2017,7:9771. |
| [37] | KWENDA S, BIRCH P R, MOLELEKI L N . Genome-wide identification of potato long intergenic noncoding RNAs responsive to Pectobacterium carotovorum subspecies brasiliense infection. BMC Genomics, 2016,17(1):614. |
| [38] | CUI J, LUAN Y, JIANG N, BAO H, MENG J . Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. The Plant Journal, 2017,89(3):577-589. |
| [39] | LI X, XING X, XU S, ZHANG M, WANG Y, WU H, SUN Z, HUO Z, CHEN F, YANG T . Genome-wide identification and functional prediction of tobacco lncRNAs responsive to root-knot nematode stress. PLoS ONE, 2018,13(11):e0204506. |
| [40] | 赵长江 . 纹枯病菌侵染后水稻防御反应相关基因的表达分析[D]. 福州: 福建农林大学, 2005. |
| ZHAO C J . Expression analysis of rice defence-related genes after infected by Rhizoctonia solani[D]. Fuzhou: Fujian Agriculture and Forestry University, 2005. (in Chinese) | |
| [41] | MALAMY J, CARR J P, KLESSIG D F, RASKIN I . Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science, 1990,250(4983):1002-1004. |
| [42] | 许志刚 . 普通植物病理学. 4版. 北京: 高等教育出版社, 2009: 296-315. |
| XU Z G . General Plant Pathology. 4th ed. Beijing: Higher Education Press, 2009: 296-315. (in Chinese) | |
| [43] | ZHU F, XI D H, YUAN S, XU F, ZHANG D W, LIN H H . Salicylic acid and jasmonic acid are essential for systemic resistance against Tobacco mosaic virus in Nicotiana benthamiana. Molecular Plant- Microbe Interactions, 2014,27(6):567-577. |
| [44] | VERBERNE M C, HOEKSTRA J, BOL J F, LINTHORST H J . Signaling of systemic acquired resistance in tobacco depends on ethylene perception. The Plant Journal, 2003,35(1):27-32. |
| [45] | BEIS K . Structural basis for the mechanism of ABC transporters. Biochemical Society Transactions, 2015,43(5):889-893. |
| [46] | HUANG T, JANDER G . Abscisic acid-regulated protein degradation causes osmotic stress-induced accumulation of branched-chain amino acids in Arabidopsis thaliana. Planta, 2017,246(4):737-747. |
| [47] | SHADLE G L, WESLEY S V, KORTH K L, CHEN F, LAMB C, DIXON R A . Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase. Phytochemistry, 2003,64(1):153-161. |
| [48] | MERCER T R, DINGER M E, MATTICK J S . Long non-coding RNAs: Insights into functions. Nature Reviews. Genetics, 2009,10(3):155-159. |
| [49] | BERTERAME N M, BERTAGNOLI S, CODAZZI V, PORRO D, BRANDUARDI P. Temperature-induced lipocalin(TIL): A shield against stress-inducing environmental shocks in Saccharomyces cerevisiae. FEMS Yeast Research, 2017, 17(6): fox056. |
| [50] | SADE D, EYBISHTZ A, GOROVITS R, SOBOL I, CZOSNEK H . A developmentally regulated lipocalin-like gene is overexpressed in Tomato yellow leaf curl virus-resistant tomato plants upon virus inoculation, and its silencing abolishes resistance. Plant Molecular Biology, 2012,80(3):273-287. |
| [1] | 裴悦宏,李凤巍,刘维娜,温玉霞,朱鑫,田绍锐,樊光进,马小舟,孙现超. 本氏烟半胱氨酸蛋白酶基因家族特征及其在TMV侵染中的功能[J]. 中国农业科学, 2022, 55(21): 4196-4210. |
| [2] | 温玉霞,张坚,王琴,王靖,裴悦宏,田绍锐,樊光进,马小舟,孙现超. 本氏烟NbMBF1c的克隆、表达及在TMV侵染过程中的功能[J]. 中国农业科学, 2022, 55(18): 3543-3555. |
| [3] | 赵珂,郑林,杜美霞,龙俊宏,何永睿,陈善春,邹修平. 柑橘SAR及其信号转导基因CsSABP2在黄龙病菌侵染中的响应特征[J]. 中国农业科学, 2021, 54(8): 1638-1652. |
| [4] | 徐翔,解屹,宋丽云,申莉莉,李莹,王勇,刘明宏,刘东阳,王小彦,赵存孝,王凤龙,杨金广. 高效靶向降解烟草花叶病毒核酸的dsRNA筛选与大量制备[J]. 中国农业科学, 2021, 54(6): 1143-1153. |
| [5] | 胡冬梅,江东,李永平,彭磊,李冬云,朱延松,杨云光. 利用Target SSR-seq技术鉴定温州蜜柑芽变材料[J]. 中国农业科学, 2021, 54(23): 5083-5096. |
| [6] | 刘昌云,李欣羽,田绍锐,王靖,裴悦宏,马小舟,樊光进,汪代斌,孙现超. 番茄SlN-like的克隆、表达与抗病毒功能[J]. 中国农业科学, 2021, 54(20): 4348-4357. |
| [7] | 陈华枝,王杰,祝智威,蒋海宾,范元婵,范小雪,万洁琦,卢家轩,郑燕珍,付中民,徐国钧,陈大福,郭睿. 蜜蜂球囊菌菌丝和孢子中长链非编码RNA的比较及潜在功能分析[J]. 中国农业科学, 2021, 54(2): 435-448. |
| [8] | 魏艳侠,李卓然,张斌,苑瑜瑾,于玮玮,常若葵,王远宏. 贝莱斯芽孢杆菌LJ02中植物免疫蛋白的筛选及其功能[J]. 中国农业科学, 2021, 54(16): 3451-3460. |
| [9] | 王继卿,郝志云,沈继源,柯娜,黄兆春,梁维炜,罗玉柱,胡江,刘秀,李少斌. 小尾寒羊泌乳性状重要lncRNAs的筛选、鉴定及功能分析[J]. 中国农业科学, 2021, 54(14): 3113-3123. |
| [10] | 周丁丁, 范元婵, 王杰, 蒋海宾, 祝智威, 范小雪, 陈华枝, 杜宇, 周紫彧, 熊翠玲, 郑燕珍, 付中民, 陈大福, 郭睿. 蜜蜂球囊菌中长链非编码RNA的调控作用[J]. 中国农业科学, 2021, 54(1): 224-238. |
| [11] | 杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿. 微小RNA及其介导的竞争性内源RNA调控网络在意大利蜜蜂工蜂中肠发育过程中的潜在作用[J]. 中国农业科学, 2020, 53(12): 2512-2526. |
| [12] | 刘培勋,万洪深,郑建敏,罗江陶,蒲宗君. 小麦PIN基因家族的鉴定及表达分析[J]. 中国农业科学, 2020, 53(12): 2321-2330. |
| [13] | 刘亦然,张虹,靳继苏,周忠实,郭建英. 莲草直胸跳甲Halloween基因家族鉴定及表达分析[J]. 中国农业科学, 2020, 53(10): 2009-2019. |
| [14] | 周丁丁,史小玉,王杰,范元婵,祝智威,蒋海宾,范小雪,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 东方蜜蜂微孢子虫孢子中长链非编码RNA的竞争性内源RNA调控网络及潜在功能[J]. 中国农业科学, 2020, 53(10): 2122-2136. |
| [15] | 汤亚飞,裴凡,李正刚,佘小漫,于琳,蓝国兵,邓铭光,何自福. 基于小RNA深度测序技术鉴定侵染广东辣椒的病毒种类[J]. 中国农业科学, 2019, 52(13): 2256-2267. |
|
||