













中国农业科学 ›› 2020, Vol. 53 ›› Issue (5): 874-889.doi: 10.3864/j.issn.0578-1752.2020.05.002
张稳,孟淑君,王琪月,万炯,马拴红,林源,丁冬( ),汤继华(
),汤继华( )
)
收稿日期:2019-07-22
接受日期:2019-09-30
出版日期:2020-03-01
发布日期:2020-03-14
通讯作者:
丁冬,汤继华
作者简介:张稳,E-mail:基金资助:
ZHANG Wen,MENG ShuJun,WANG QiYue,WAN Jiong,MA ShuanHong,LIN Yuan,DING Dong( ),TANG JiHua(
),TANG JiHua( )
)
Received:2019-07-22
Accepted:2019-09-30
Online:2020-03-01
Published:2020-03-14
Contact:
Dong DING,JiHua TANG
摘要:
【目的】叶绿素是参与光合途径最为重要的光合色素。叶绿体的发育及叶绿素的合成在很大程度上依赖于质体基因组与核基因组之间的双向信号传导来精确协调基因表达。通过对白化表型的CRISPR/Cas9-ZmpTAC2转基因阳性纯合突变材料进行RNA-seq研究,筛选和鉴定参与叶绿素合成的相关基因,为明确叶绿素的合成途径奠定基础。【方法】以CRISPR/Cas9-ZmpTAC2玉米转基因编辑纯合突变株系为研究材料,使用透射电镜观察叶绿体超微结构和分光光度法测定叶片叶绿素含量,确定叶绿体发育状态及叶绿素合成情况。对转基因阴性材料(CK)和CRISPR/Cas9-ZmpTAC2转基因纯合编辑材料(zmptac2)苗期叶片取样进行转录组测序。通过生物信息学分析,寻找CK与zmptac2间差异表达的基因;qRT-PCR对差异表达基因进行验证。通过酵母双杂交筛选与玉米pTAC2互作的蛋白质。【结果】共获得15株T0转基因植株,包括绿色植株(7株)和白色植株(8株)。绿色幼苗中3株为转基因阴性材料,4株为转基因阳性(2株为未编辑,2株为杂合编辑突变),白色植株(8株)均为转基因阳性纯合编辑。与CK相比,突变体(zmptac2)叶绿体发异常,叶绿素含量显著降低。RNA-seq的结果显示,CK与zmptac2之间共检测到1 367个基因差异表达,其中618个基因上调表达(zmptac2/CK),749个基因下调表达(zmptac2/CK)。GO富集分析显示,下调基因显著富集到叶绿体和质体中。KEGG分析表明下调表达基因显著富集在苯丙氨酸代谢、酪氨酸代谢和异喹啉生物碱生物合成等途径。选取的15个差异基因表达模式均与测序数据相一致,表明测序结果是可靠的。与CK相比,zmptac2中依赖PEP(plastid-encoded RNA polymerase)转录的基因表达量显著降低,而依赖NEP(nuclear gene-encoded RNA polymerase)转录的基因表达量则显著上升。通过对玉米cDNA文库筛选和互作验证,鉴定出ZmpTAC3与ZmpTAC2存在互作。【结论】ZmpTAC2突变会导致叶绿体早期生物合成受阻,该基因参与叶绿体发育及叶绿素合成,且该种作用是由ZmpTAC2调控PEP相关基因表达而实现的。
张稳,孟淑君,王琪月,万炯,马拴红,林源,丁冬,汤继华. 玉米pTAC2影响苗期叶片叶绿素合成的转录组分析[J]. 中国农业科学, 2020, 53(5): 874-889.
ZHANG Wen,MENG ShuJun,WANG QiYue,WAN Jiong,MA ShuanHong,LIN Yuan,DING Dong,TANG JiHua. Transcriptome Analysis of Maize pTAC2 Effects on Chlorophyll Synthesis in Seedling Leaves[J]. Scientia Agricultura Sinica, 2020, 53(5): 874-889.
表1
CRISPR/Cas9系统潜在脱靶位点突变检测"
| 编号 Serial number | 潜在脱靶位置 Putative off-target locus | 潜在脱靶序列 Sequence of the putative off-target site (5′-3′) | 错配碱基数 No. of mismatching bases | 测试株数 T0 No. of plants tested | 突变株数 T0 No. of plants with mutations |
|---|---|---|---|---|---|
| Off-target1 | Chr.7:109756398-109756420 | AgCTGGaGTAtAGCCTtATACGG | 4 | 8 | 0 |
| Off-target2 | Chr.1 :145160658-145160680 | AcCTGGGGTACAGCCaGATcAGG | 3 | 8 | 0 |
| Off-target3 | Chr.6:44283532-44283554 | cGaCCACGCAGCTATGTcTGAGG | 3 | 8 | 0 |
| Off-target4 | Chr.4:230687431-230687453 | cGaCCACGCAGCTATGTcTGAGG | 3 | 8 | 0 |
| Off-target5 | Chr.7:8936191-8936213 | TGGCCACGCgGCTAcGTgTGGGG | 3 | 8 | 0 |
| Off-target6 | Chr.7:8936289-8936311 | TGGCCACGCgGCTAcGTgTGGGG | 3 | 8 | 0 |
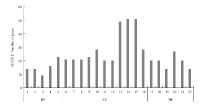
图3
差异表达基因GO富集分析 1:细胞或亚细胞成分的运动Movement of cell or subcellular component;2:基于微管的运动Microtubule-based movement;3:有丝分裂细胞周期的调控Regulation of mitotic cell cycle;4:调节细胞成分的组织Regulation of cellular component organization;5:微管过程Microtubule-based process;6:核小体Nucleosome;7:蛋白质-DNA复合物Protein-DNA complex;8:DNA包装复合体DNA packaging complex;9:染色质Chromatin;10:染色体部分Chromosomal part;11:叶绿体基质Chloroplast stroma;12:质体基质Plastid stroma;13:细胞外区域Extracellular region;14:叶绿体Chloroplast;15:质体Plastid;16:染色体Chromosome;17:微管结合Microtubule binding;18:微管蛋白结合Tubulin binding;19:微管运动活动Microtubule motor activity;20:细胞骨架蛋白结合Cytoskeletal protein binding;21:蛋白质异二聚活性Protein heterodimerization activity;22:运动活动Motor activity。BP:生物学过程 Biological process;CC:细胞组分Cellular component;MF:分子功能Molecular function;差异表达基因GO注释,横坐标为GO功能分类,纵坐标为注释到GO条目上的基因数The differentially expressed gene GO annotation, the abscissa is the GO functional classification, and the ordinate is the number of genes annotated to the GO term"
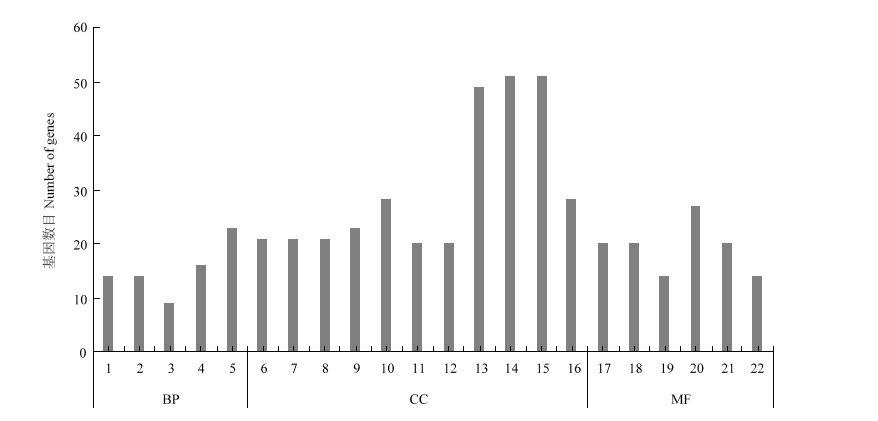
表3
qRT-PCR验证的差异表达基因"
| 基因编号 Gene ID | zmptac2 | CK | 差异倍数 log2Fold change | P值 P-value | 基因名称 Synonyms | 描述 Description |
|---|---|---|---|---|---|---|
| Zm00001d039206 | 189.7162285 | 4.75059079 | 5.370830095 | 7.82E-06 | Bzip118 | bZIP转录因子超家族蛋白 Putative bZIP transcription factor superfamily protein |
| Zm00001d043825 | 80.63331881 | 0 | 8.789501589 | 0.00185833 | CtpA1 | 羧基末端加工肽酶1 Carboxyl-terminal-processing peptidase 1 chloroplastic |
| Zm00001d027476 | 176.7085969 | 0 | 9.921454443 | 4.88E-06 | DNA polymerases | 依赖DNA的DNA聚合酶 DNA-directed DNA polymerases |
| Zm00001d031503 | 1000.059518 | 22.40077408 | 5.473911936 | 0.00680168 | CPN60 | 伴侣蛋白亚基 Chaperonin 60 subunit alpha 2 chloroplastic |
| Zm00001d008221 | 0 | 351.1761292 | -21.80949 | 1.54E-09 | Cyc5 | 细胞周期蛋白5 Cyclin5 |
| Zm00001d022599 | 363.2261699 | 3395.326772 | -3.03756614 | 5.64E-14 | ZmpTAC2 | 质体转录活性染色体2 Plastid transcriptionally active chromosome 2 homolog |
| Zm00001d033365 | 0 | 362.4571584 | -23.97922 | 0.0000335 | Cyclin A3 1 | 细胞周期蛋白A3-1 Putative cyclin-A3-1 |
| Zm00001d027540 | 0 | 1170.714751 | -25.15007 | 0.0000113 | Gst12 | 谷胱甘肽S-转移酶12 Glutathione S-transferase12 |
| Zm00001d048353 | 0 | 168.1203061 | -22.56015 | 1.08E-08 | Gst13 | 谷胱甘肽转移酶13 Glutathione transferase13 |
| Zm00001d027760 | 0 | 1660.523115 | -24.97806 | 1.32E-05 | H2A | 组蛋白H2A Histone H2A |
| Zm00001d051591 | 10.74539904 | 2639.275824 | -7.938035 | 4.88E-05 | his2b5 | 组蛋白2B5 Histone 2B5 |
| Zm00001d042730 | 87.36474604 | 4052.072328 | -5.535586 | 5.51E-05 | H3 | 组蛋白H3 Histone H3 |
| Zm00001d012276 | 103.9578986 | 6414.512969 | -5.947576 | 0.000341 | H3.2 | 组蛋白H3.2 Histone H3.2 |
| Zm00001d039497 | 40.62122946 | 4971.062871 | -6.93312404 | 8.97E-28 | ClpC1 | 伴侣蛋白ClpC1(线粒体) Chaperone protein ClpC1 chloroplastic |
| Zm00001d042062 | 0 | 2272.297402 | -9.395263783 | 0.00021753 | ORG3 | 转录因子ORG3 Transcription factor ORG3 |
表4
酵母双杂筛库获得ZmpTAC2的互作蛋白质"
| 编号 No. | 基因编号 Gene ID_v4 | 亚细胞定位 Subcellular localization | 基因描述 Gene description |
|---|---|---|---|
| 1 | Zm00001d022242 | 细胞膜Cell membrane | 胞间连丝胼胝质结合蛋白5 PLASMODESMATA CALLOSE-BINDING PROTEIN 5 |
| 2 | Zm00001d010039 | 细胞壁Cell wall | 含纤维素酶的蛋白质Cellulase containing protein |
| 3 | Zm00001d023305 | 叶绿体Chloroplast | Protein LURP-one-related 2 |
| 4 | Zm00001d049153 | 叶绿体Chloroplast | 催化/水解酶Catalytic/ hydrolase |
| 5 | Zm00001d041550 | 叶绿体Chloroplast | 热休克蛋白3 Heat shock 70 kDa protein 3 |
| 6 | Zm00001d035270 | 叶绿体Chloroplast | SAM结构域蛋白Sterile alpha motif (SAM) domain-containing protein |
| 7 | Zm00001d028221 | 叶绿体Chloroplast | ATP合酶4 ATP synthase4 |
| 8 | Zm00001d046073 | 叶绿体Chloroplast | Protein trichome birefringence-like 26 |
| 9 | Zm00001d032373 | 叶绿体Chloroplast | 叶绿体RNA剪接体1 Chloroplast RNA splicing1 |
| 10 | Zm00001d007417 | 叶绿体Chloroplast | ATP依赖性DNA解旋酶ATP-dependent DNA helicase chloroplastic |
| 11 | Zm00001d024248 | 叶绿体Chloroplast | PPR蛋白Pentatricopeptide repeat*-59040 |
| 12 | Zm00001d016660 | 细胞质Cytoplasm | 肽基脯氨酰异构酶1 Peptidyl-prolyl isomerase1 |
| 13 | Zm00001d033623 | 细胞质Cytoplasm | 脂氧合酶 Lipoxygenase |
| 14 | Zm00001d018441 | 细胞质,细胞核 Cytoplasm. Nucleus | 核苷三磷酸水解酶超家族蛋白 P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| 15 | Zm00001d040681 | 细胞质,细胞核 Cytoplasm. Nucleus | alpha/beta水解超家族蛋白 Alpha/beta-Hydrolases superfamily protein |
| 16 | Zm00001d038857 | 线粒体Mitochondrion | 伴侣蛋白CPN60 Chaperonin CPN60-like 2 mitochondrial |
| 17 | Zm00001d035201 | 线粒体Mitochondrion | 60S核糖体蛋白P060S acidic ribosomal protein P0 |
| 18 | Zm00001d053434 | 细胞核Nucleus | RING锌指结构域超家族蛋白 Putative RING zinc finger domain superfamily protein |
| 19 | Zm00001d032422 | 细胞核Nucleus | 休眠相关蛋白1 Dormancy-associated protein 1 |
| 20 | Zm00001d044246 | 细胞核Nucleus | 组蛋白H2A Histone H2A |
| 21 | Zm00001d044429 | 细胞核Nucleus | 硫氧还蛋白超家族蛋白 Thioredoxin superfamily protein |
| 22 | Zm00001d021954 | 细胞核Nucleus | MO25蛋白 Putative MO25-like protein |
| 23 | Zm00001d032646 | 细胞核Nucleus | RHC1A |
| 24 | Zm00001d016876 | 细胞核Nucleus | Trihelix转录因子ASIL2 Trihelix transcription factor ASIL2 |
| 25 | Zm00001d043914 | 细胞核Nucleus | Homeobox-DDT结构域蛋白RLT1 Homeobox-DDT domain protein RLT1 |
| 26 | Zm00001d014858 | 细胞核Nucleus | DNA结合蛋白MNB1B DNA-binding protein MNB1B |
| 27 | Zm00001d036494 | 细胞核Nucleus | GATA转录因子25 GATA transcription factor 25 |
| 28 | Zm00001d013159 | 细胞核Nucleus | α-微管蛋白3 Alpha-tubulin 3 |
| 29 | Zm00001d001847 | 细胞核Nucleus | NUMOD3基序家族蛋白 NUMOD3 motif family protein expressed; protein |
| 30 | Zm00001d032790 | 细胞核Nucleus | 质体转录活性3 Plastid transcriptionally active 3 |
| 31 | Zm00001d052298 | 细胞核Nucleus | 锌指(C3HC4型RING指)家族蛋白 Zinc finger (C3HC4-type RING finger) family protein |
| 32 | Zm00001d027652 | 液泡Vacuole | 水通道蛋白TIP1-1 Aquaporin TIP1-1 |
| [1] | 刘红梅, 周新跃, 刘建丰, 邱颖波, 范峰峰, 徐庆国 . 籼型杂交稻光合特性的配合力分析. 植物遗传资源学报, 2014,15(4):699-705. |
| LIU H M, ZHOU X Y, LIU J F, QIU Y B, FAN F F, XU Q G . Analysis of combining ability of photosynthetic characteristics in Indica hybrid rice. Journal of Plant Genetic Resources, 2014,15(4):699-705. (in Chinese) | |
| [2] | 李合生 . 现代植物生理学. 北京: 高等教育出版社, 2012. |
| LI H S . Modern Plant Physiology. Beijing: Higher Education Press, 2012. ( in Chinese) | |
| [3] | LEISTER D . Chloroplast research in the genomic age. Trends in Genetics, 2003,19(1):47-56. |
| [4] | SUGIMOTO H, KUSUMI K, TOZAWA Y, YAZAKI J, KISHIMOTO N, KIKUCHI S, IBA K . The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant and Cell Physiology, 2004,45(8):985-996. |
| [5] | ABDALLAH F, SALAMINI F, LEISTER D . A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends in Plant Science, 2000,5(4):141-142. |
| [6] | PELTIER J B, CAI Y, SUN Q, ZABROUSKOV V, GIACOMELLI L, RUDELLA A, YTTERBERG A J, RUTSCHOW H, VAN WIJK K J . The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Molecular & Cellular Proteomics, 2006,5(1):114-133. |
| [7] | BÖRNER T, ALEYNIKOVA A Y, ZUBO Y O, KUSNETSOV V V . Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochimica et Biophysica Acta, 2015,1847(9):761-769. |
| [8] | SHIINA T, TSUNOYAMA Y, NAKAHIRA Y, KHAN M S . Plastid RNA polymerases, promoters, and transcription regulators in higher plants. International Review of Cytology-a Survey of Cell Biology, 2005,244:1-68. |
| [9] | 叶琳珊 . pTAC7调控拟南芥叶绿基因表达的机理研究[D]. 上海: 上海师范大学, 2015. |
| YE L S . Mechanism of pTAC7 regulation of Arabidopsis leaf green gene expression[D]. Shanghai: Shanghai Normal University, 2015. ( in Chinese) | |
| [10] | 王振国, 魏家绵, 沈允钢 . 玉米叶绿体atpB及atpE基因在大肠杆菌中的表达. 植物生理学报, 1995(3):265-396. |
| WANG Z G, WEI J M, SHEN Y G . Over-expression of maize chloroplastsatpB and atpE genes in Escherichia coli. Journal of Plant Physiology, 1995(3):265-396. (in Chinese) | |
| [11] | PUTHIYAVEETIL S, IBRAHIM I M, JELICIĆ B, TOMASIĆ A, FULGOSI H, ALLEN J F . Transcriptional control of photosynthesis genes: The evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biology and Evolution Biology and Evolution, 2010,2:888-896. |
| [12] | ZHELYAZKOVA P, SHARMA C M, FÖRSTNER K U, LIERE K, VOGEL J, BÖRNER T . The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. The Plant Cell, 2012,24:123-136. |
| [13] | LGLOI G L, KÖSSEL H . The transcriptional apparatus of chloroplasts. Critical Reviews in Plant Sciences, 1992,10(6):525-558. |
| [14] | KRAUSE K, MAIER R M, KOFER W, KRUPINSKA K, HERRMANN R G . Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Molecular and General Genetics, 2000,263:1022-1030. |
| [15] | PFALZ J, LIERE K, KANDLBINDER A, DIETZ K J, OELMULLER R . pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. The Plant Cell, 2006,18(1):176-197. |
| [16] | CHEN M, GALVAO R M, LI M, BURGER B, BUGEA J, BOLADO J, CHORY J . Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell, 2010,141:1230-1240. |
| [17] | MYOUGA F, HOSODA C, UMEZAWA T, IIZUMI H, KUROMORI T, MOTOHASHI R, SHONO Y, NAGATA N, IKEUCHI M, SHINOZAKI K . A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. The Plant Cell, 2008,20:3148-3162. |
| [18] | KROLL D, MEIERHOFF K, BECHTOLD N, KINOSHITA M, WESTPHAL S, VOTHKNECHT U C, SOLL J, WESTHOFF P . VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proceedings of the National Academy of Sciences of the United States of America, 2001,98:4238-4242. |
| [19] | MARÉCHAL A, PARENT J S, VÉRONNEAU-LAFORTUNE F, JOYEUX A, LANG B F, BRISSON N . Whirly proteins maintain plastid genome stability in Arabidopsis. Proceedings of the National Academy of Sciences of the USA, 2009,106(34):14693-14698. |
| [20] | GAO Z P, YU Q B, ZHAO T T, MA Q, CHEN G X, YANG Z N . A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiology, 2011,157(4):1733-1745. |
| [21] | YAGI Y, ISHIZAKI Y, NAKAHIRA Y, TOZAWA Y, SHIINA T . Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proceedings of the National Academy of Sciences of the USA, 2012,109(19):7541-7546. |
| [22] | YU Q B, LU Y, ZHAO T T, HUANG C, ZHAO H F, ZHANG X L. LV R H, YANG Z N . TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiologia Plantarum, 2013,148(3):408-421. |
| [23] | 张森 . 果糖激酶类似蛋白1,2与硫氧还蛋白Z互作调控水稻叶绿体发育[D]. 北京: 中国农业科学院, 2018. |
| ZHANG S . Fructose kinase-like protein 1,2 interacts with thioredoxin Z to regulate rice chloroplast development[D]. Beijing: Chinese Academy of Agricultural Sciences, 2018. ( in Chinese) | |
| [24] | KUSABA M, ITO H, MORIT R, IIDA S, SATO Y, FUJIMOTO M, KAWASAKI S, TANAKA R, HIROCHIKA H, NISHIMURA M . Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. The Plant Cell, 2007,19(4):1362-1375. |
| [25] | TANG Y Y, LI M, CHEN Y P, WU P Z, WU G J, JIANG H W . Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. Journal of Plant Physiology, 2011,168(16):1952-1959. |
| [26] | HAYASHI-TSUGANE M, TAKAHARA H, AHMED N, HIMI E, TAKAGI K, IIDA S, TSUGANE K, MAEKAWA M . A mutable albino allele in rice reveals that formation of thylakoid membranes requires the SNOW-WHITE LEAF1 gene. Plant & Cell Physiology, 2014,55(1):3-15. |
| [27] | ZHAO D S, ZHANG C Q, LI Q F, YANG Q Q, GU M H, LIU Q Q . A residue substitution in the plastid ribosomal protein L12/AL1 produces defective plastid ribosome and causes early seedling lethality in rice. Plant Molecular Biology, 2016,91(1/2):161-177. |
| [28] | WANG D, LIU H G, ZHAI G W, WANG L S, SHAO J F, TAO Y Z . Osptac2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. Journal of Genetics & Genomics, 2016,43(10):601-608. |
| [29] | 胡根海, 喻树迅 . 利用改良的CTAB法提取棉花叶片总RNA. 棉花学报, 2007(1):69-70. |
| HU G H, YU S X . Extraction of total RNA from cotton leaves by improved CTAB method. Cotton Science, 2007(1):69-70. (in Chinese) | |
| [30] | 刘仁林, 虞功清, 邹伟民, 杜天真 . 植物透射电镜样品制备技术的改进. 江西林业科技, 2008(1):43-45. |
| LIU R L, YU G Q, ZOU W M, DU T Z . Improvements in technique of madding ultrathin section for transmission electron microscope. Jiangxi Forestry Science and Technology, 2008(1):43-45. (in Chinese) | |
| [31] | KWON K, CHO M . Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. The Plant Journal, 2008,55(3):428-442 |
| [32] | ANDREWS S . FastQC: A quality control tool for high throughput sequence data. 2013. |
| [33] | BOLGER A M, LOHSE M, USADEL B . Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 2014,30(15):2114-2120. |
| [34] | KIM D, LANGMEAD B, SALZBERG S L . HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 2015,12(4):357-360. |
| [35] | LIAO Y, SMYTH G K, SHI W . Feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England), 2014,30(7):923-930. |
| [36] | YU G, WANG L G, HAN Y, HE Q Y . Cluster Profiler: An R package for comparing biological themes among gene clusters. A Journal of Integrative Biology, 2012,16(5):284-287. |
| [37] | LIVAK K J, SCHMITTGEN T D . Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method . Methods, 2001,25(4):402-408. |
| [38] | YE R, YAO Q H, XU Z H, WEI X H . Development of an efficient method for the isolation of factors involved in gene transcription during rice embryo development. Plant Journal, 2004,38(2):348-357. |
| [39] | MEINKE D, MURALLA R, SWEENEY C, DICKERMAN A . Identifying essential genes in Arabidopsis thaliana. Trends in Plant Science, 2008,13(9):483-491. |
| [40] | WATERS M T, LANGDALE J A . The making of a chloroplast. The EMBO Journal, 2014,28(19):2861-2873. |
| [41] | 吴久利 . 两种氨基酸和三种遗传病. 生物学教学, 2005,30(10):72. |
| WU J L . Two kinds of amino acids and three genetic diseases. Biology Teaching, 2005,30(10):72. (in Chinese) | |
| [42] | NAKAMURA H, MURAMATSU M, HAKATA M, UENO O, NAGAMURA Y, HIROCHIKA H, TAKANO M, ICHIKAWA H . Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant and Cell Physiology, 2009,50(11):1933-1949. |
| [43] | SCHARFENBERG M, MITTERMAYR L, VON ROEPENACK- LAHAYE E, SCHLICKE H, GRIMM B, LEISTER D, KLEINE T. Functional characterization of the two ferrochelatasein Arabidopsis thaliana. Plant Cell and Environment, 2013,38(2):280-298. |
| [44] | SAHA A, DAS S, MOIN M, DUTTA M, BAKSHI A, MADHAV M S, KIRTI P B . Genome-wide identification and comprehensive expression profiling of ribosomal protein small subunit (RPS) genes and their comparative analysis with the large subunit (RPL) genes in rice. Frontiers in Plant Science, 2017,8:1553-1574. |
| [45] | 张丽君 . 蕨类植物叶绿体rps4基因和PHY基因GAF结构域的适应性进化研究[D]. 北京:中国科学院大学, 2010. |
| ZHANG L J . Adaptive evolution of chlorophyll rps4 gene and PHY gene GAF domain in ferns[D]. Beijing: University of Chinese Academy of Sciences, 2010. ( in Chinese) | |
| [46] | OELMÜLLER R, HERRMANN R G, PAKRASI H B . Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. Journal of Biological Chemistry, 1996,271(36):21848-21852. |
| [47] | 郑唐春 . 小黑杨PsnCYCD1;1基因参与细胞分裂及次生生长作用机理研究[D]. 哈尔滨:东北林业大学, 2016. |
| ZHENG T C . Mechanism of PsnCYCD1;1 gene involved in cell division and secondary growth[D]. Harbin: Northeast Forestry University, 2016. ( in Chinese) | |
| [48] | 梁彬, 邹向阳 . 细胞周期蛋白依赖性激酶及抑制因子与肿瘤. 中国实验诊断学, 2006,10(11):1378-1380. |
| LIANG B, ZOU X Y . Cyclin-dependent kinases and inhibitors and tumors. Chinese Journal of Laboratory Diagnosis, 2006,10(11):1378-1380. (in Chinese) | |
| [49] | 赵春玲, 宋咏梅, 樊飞跃, 詹启敏 . 细胞周期蛋白cyclin b1与肿瘤. 肿瘤, 2007,27(4):322-326. |
| ZHAO C L, SONG Y M, FAN F Y, ZHAN Q M . Cyclin cyclin b1 and tumor. Tumor, 2007,27(4):322-326. | |
| [50] | VANNESTE S, COPPENS F, LEE E, DONNER T J, XIE Z, VAN ISTERDAEL G, DHONDT S, DE WINTER F, DE RYBEL B, VUYLSTEKE M, DE VEYLDER L, FRIML J, INZÉ D, GROTEWOLD E, SCARPELLA E, SACK F, BEEMSTER G T, BEECKMAN T . Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. The EMBO Journal, 2011,30(16):3430-3441. |
| [51] | RUDELLA A, FRISO G, ALONSO M, ECKER J, WIJK K . Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. The Plant Cell, 2006,18(7):1704-1721. |
| [52] | NISHIMURA K, ASAKURA Y, FRISO G, KIM J, OH S H, RUTSCHOW H, PONNALA L, VAN WIJK K J . ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. The Plant Cell, 2013,25(6):2276-2301. |
| [53] | SJOGREN L L, MACDONALD T M, SUTINEN S, CLARKE A K . Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiology, 2004,136:4114-4126. |
| [1] | 赵政鑫,王晓云,田雅洁,王锐,彭青,蔡焕杰. 未来气候条件下秸秆还田和氮肥种类对夏玉米产量及土壤氨挥发的影响[J]. 中国农业科学, 2023, 56(1): 104-117. |
| [2] | 柴海燕,贾娇,白雪,孟玲敏,张伟,金嵘,吴宏斌,苏前富. 吉林省玉米穗腐病致病镰孢菌的鉴定与部分菌株对杀菌剂的敏感性[J]. 中国农业科学, 2023, 56(1): 64-78. |
| [3] | 李周帅,董远,李婷,冯志前,段迎新,杨明羡,徐淑兔,张兴华,薛吉全. 基于杂交种群体的玉米产量及其配合力的全基因组关联分析[J]. 中国农业科学, 2022, 55(9): 1695-1709. |
| [4] | 熊伟仡,徐开未,刘明鹏,肖华,裴丽珍,彭丹丹,陈远学. 不同氮用量对四川春玉米光合特性、氮利用效率及产量的影响[J]. 中国农业科学, 2022, 55(9): 1735-1748. |
| [5] | 李易玲,彭西红,陈平,杜青,任俊波,杨雪丽,雷鹿,雍太文,杨文钰. 减量施氮对套作玉米大豆叶片持绿、光合特性和系统产量的影响[J]. 中国农业科学, 2022, 55(9): 1749-1762. |
| [6] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [7] | 马小艳,杨瑜,黄冬琳,王朝辉,高亚军,李永刚,吕辉. 小麦化肥减施与不同轮作方式的周年养分平衡及经济效益分析[J]. 中国农业科学, 2022, 55(8): 1589-1603. |
| [8] | 李前,秦裕波,尹彩侠,孔丽丽,王蒙,侯云鹏,孙博,赵胤凯,徐晨,刘志全. 滴灌施肥模式对玉米产量、养分吸收及经济效益的影响[J]. 中国农业科学, 2022, 55(8): 1604-1616. |
| [9] | 张家桦,杨恒山,张玉芹,李从锋,张瑞富,邰继承,周阳晨. 不同滴灌模式对东北春播玉米籽粒淀粉积累及淀粉相关酶活性的影响[J]. 中国农业科学, 2022, 55(7): 1332-1345. |
| [10] | 谭先明,张佳伟,王仲林,谌俊旭,杨峰,杨文钰. 基于PLS的不同水氮条件下带状套作玉米产量预测[J]. 中国农业科学, 2022, 55(6): 1127-1138. |
| [11] | 赵凌, 张勇, 魏晓东, 梁文化, 赵春芳, 周丽慧, 姚姝, 王才林, 张亚东. 利用高密度Bin图谱定位水稻抽穗期剑叶叶绿素含量QTL[J]. 中国农业科学, 2022, 55(5): 825-836. |
| [12] | 冯宣军, 潘立腾, 熊浩, 汪青军, 李静威, 张雪梅, 胡尔良, 林海建, 郑洪建, 卢艳丽. 南方地区120份甜、糯玉米自交系重要目标性状和育种潜力分析[J]. 中国农业科学, 2022, 55(5): 856-873. |
| [13] | 刘苗,刘朋召,师祖姣,王小利,王瑞,李军. 氮磷配施下夏玉米临界氮浓度稀释曲线的构建与氮营养诊断[J]. 中国农业科学, 2022, 55(5): 932-947. |
| [14] | 乔远,杨欢,雒金麟,汪思娴,梁蓝月,陈新平,张务帅. 西北地区玉米生产投入及生态环境风险评价[J]. 中国农业科学, 2022, 55(5): 962-976. |
| [15] | 黄兆福, 李璐璐, 侯梁宇, 高尚, 明博, 谢瑞芝, 侯鹏, 王克如, 薛军, 李少昆. 不同种植区玉米生理成熟后田间站秆脱水的积温需求[J]. 中国农业科学, 2022, 55(4): 680-691. |
|
||