













中国农业科学 ›› 2020, Vol. 53 ›› Issue (5): 857-873.doi: 10.3864/j.issn.0578-1752.2020.05.001
收稿日期:2019-05-21
接受日期:2019-09-30
出版日期:2020-03-01
发布日期:2020-03-14
通讯作者:
刘旭
作者简介:宋松泉,E-mail:sqsong@ibcas.ac.cn。
基金资助:
SONG SongQuan1,4,LIU Jun2,XU HengHeng2,LIU Xu3( ),HUANG Hui4
),HUANG Hui4
Received:2019-05-21
Accepted:2019-09-30
Online:2020-03-01
Published:2020-03-14
Contact:
Xu LIU
摘要:
种子休眠是许多植物在长期系统发育进程中获得的一种适应环境变化的特性,是调控种子萌发和幼苗形成的最适时空分布的一种有效方式,也是物种成功繁衍与传播的一种选择性策略。种子休眠与萌发的激素调控可能是一种高度保守的机制,其中脱落酸(ABA)在种子休眠解除与萌发中起关键作用,赤霉素(GA)在休眠被解除后促进种子萌发。ABA在种子休眠与萌发中的作用主要受ABA代谢(生物合成和分解代谢)和信号传递途径的调控。为此,本文在综述ABA代谢和信号传递研究进展的基础上,阐述了ABA在种子发育、休眠与萌发中的作用,以及种子休眠特异性基因DOG1(萌发延迟1)与ABA信号组分的关系。研究表明,C40环氧类胡萝卜素是ABA生物合成的前体,玉米黄质环氧化酶和9-顺式-环氧类胡萝卜素二加氧酶是ABA生物合成的主要调节酶;ABA的分解代谢包括羟基化作用和与葡萄糖结合,CYP707A家族催化ABA C-8'位置上的羟基化作用,这是ABA分解代谢的重要步骤。在核心ABA信号传递途径中,ABA与PYR/PYL/RCAR受体结合并触发受体发生构象变化,从而允许受体-ABA复合物与2C类蛋白磷酸酶(PP2C)结合并抑制其活性,导致激酶如蔗糖非发酵-1相关的蛋白激酶2(SnRK2)的去抑制和活化。然后,这些激酶磷酸化和活化转录因子(transcription factors,TF),TF与靶启动子结合和诱导下游的ABA反应基因表达。ABA在种子成熟中后期积累,合子组织中合成的ABA诱导初生休眠和促进种子成熟;在发育中积累和在干种子中存留的ABA含量在种子吸胀初期下降。ABA是种子休眠诱导和维持的正调控因子,是萌发的负调控因子。DOG1在种子成熟过程中表达和发挥作用,其表达受可变剪接和可变多腺苷酸化调控。反义DOG1是种子休眠的一种抑制因子,通过干扰转录和转录延伸负调控DOG1的表达和种子休眠。种子的休眠与萌发除了被核心ABA信号途径调控外,也被DOG1-AHG1(ABA过敏感萌发1)/AHG3途径调控。DOG1能与AHG1/AHG3结合,通过结合ABA信号传递的负调控因子和增加对ABA的敏感性而引起种子休眠。最后,提出了该领域需要进一步研究的科学问题,包括ABA代谢中ABA 8'-羟化酶、ABA葡糖基转移酶和β-葡糖苷酶及其基因怎样响应发育和环境的变化以维持正常的ABA水平。ABA的重要调控因子例如Ca 2+或者活性氧对核心ABA信号传递途径的影响,核心ABA信号传递途径与DOG1-AHG1/AHG3途径的下游重叠组分PP2C在整合生理条件或者环境信号时优先响应哪一条途径、这两条途径怎样被协调、以及PP2C有哪些新的靶组分。本文将为深入研究ABA调控种子休眠与萌发的分子机理提供参考。
宋松泉,刘军,徐恒恒,刘旭,黄荟. 脱落酸代谢与信号传递及其调控种子休眠与萌发的分子机制[J]. 中国农业科学, 2020, 53(5): 857-873.
SONG SongQuan,LIU Jun,XU HengHeng,LIU Xu,HUANG Hui. ABA Metabolism and Signaling and Their Molecular Mechanism Regulating Seed Dormancy and Germination[J]. Scientia Agricultura Sinica, 2020, 53(5): 857-873.
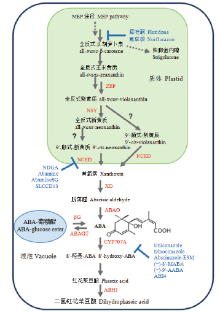
图1
ABA生物合成和分解代谢途径(根据DEJONGHE等[13]修改) ABA前体是由甲基赤藓糖醇磷酸(MEP)途径合成的。酶用红色表示。ZEP:玉米黄质环氧化酶;NSY:新黄质合酶;NCED:9-顺式-环氧类胡萝卜素双加氧酶;XD:黄氧素脱氢酶;ABAO:脱落醛氧化酶;CYP707A:ABA 8'-羟化酶;ABH1:红花菜豆酸还原酶1;ABAGT:ABA葡糖基转移酶;βG:β-葡糖苷酶。酶的抑制剂用蓝色表示。(+)-9'-AABA:(+)-9'-乙炔-ABA;AHI4:ABA 8'-羟化酶抑制剂4;(+)-8'-MABA:(+)-8'-次甲基-ABA;NDGA:去甲二氢愈创木酸;SLCCD13:类倍半萜类胡萝卜素裂解双加氧酶抑制剂13"
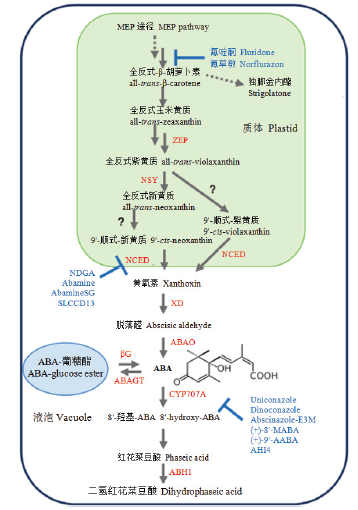
图3
ABA信号传递途径和DOG1调控种子休眠的新模型(根据NONOGAKI[19]修改) 在ABA感受和信号传递途径中(左),ABA受体(PYR/PYL/RCAR)与ABA不敏感1(ABI1)亚家族2C类蛋白磷酸酶(PP2C)包括ABI1、ABI2、ABA过敏感1(HAB1)和HAB2结合,并使PP2C失活,从而导致激酶例如蔗糖非发酵-1相关的蛋白激酶2(SnRK2)的去抑制和活化。这些激酶然后磷酸化和活化转录因子(TF),TF与靶启动子(Pro)结合,诱导下游的ABA反应基因。对于种子休眠的调节(右),DOG1与ABA过敏感萌发1(AHG1)和AHG3结合,PP2C主要在种子中起作用。DOG1被认为是通过束缚这些ABA信号传递的负调控因子和增加种子对ABA的敏感性而引起种子休眠"
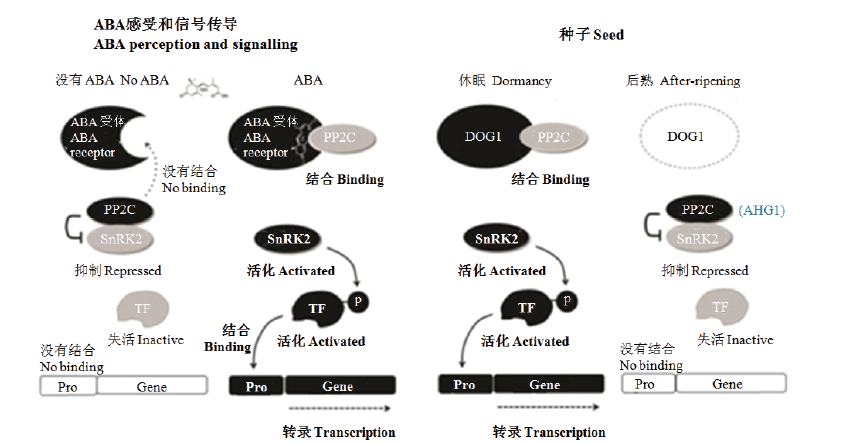
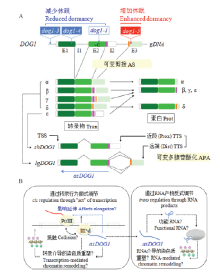
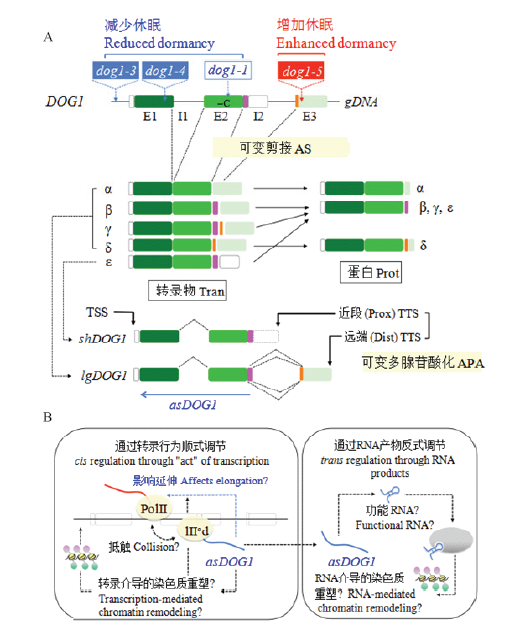
图4
DOG1表达和功能的调节(引自NONOGAKI[91]) A:DOG1的结构。顶部:具有外显子(E1、E2、E3)和内含子(I1、I2)的DOG1基因组DNA。可变剪接区域用粉红色和橙色作标记。表明dog1突变(dog1-3、dog1-4和dog1-5中的T-DNA,以及dog1-1中的单个碱基缺失(-C))的大致位置。中部:可变的DOG1转录物(α、β、γ、δ、ε)和相应的蛋白。注意DOG1-ε不是一个真正意义上的可变剪接产物。底部:可变多腺苷酸化的短DOG1(shDOG1,与DOG1-ε相同)和长DOG1(lgDOG1,包括DOG1-α、DOG1-β、DOG1-γ和DOG1-δ)转录物。转录起始(TSS)和终止(TTS)位点被表明。反义DOG1(asDOG1)的大致位置和方向用蓝色箭头标明。B:AsDOG1功能的可能机制。相对稳定的asDOG1 RNA可能以一种序列专一的方式或者通过它的二级结构作为一种调节RNA起作用,用于RNA介导的染色质重塑(右图,反式调节)。然而,等位基因专一的asDOG1的表达已经表明asDOG1在顺式调节中起作用(左图)。转录本身的“行为”而不是转录产物(RNA)发挥asDOG1的表达对DOG1表达和休眠的负面作用。反义表达可能引起转录干扰和影响转录延伸,这对DOG1表达和种子休眠是重要的;而转录介导的染色质重塑也是可能的。AS:可变剪接;APA:可变多腺苷酸化;Dist:远端;Prox:近段;Prot:蛋白;Tran:转录物"
| [1] | BEWLEY J D, BRADFORD K J , HILHORST H W M , NONOGAKI H . Physiology of Development, Germination and Dormancy. 3rd ed. New York: Springer, 2013. |
| [2] | 邓志军, 宋松泉, 艾训儒, 姚兰 . 植物种子保存和检测的原理与技术. 北京: 科学出版社, 2019. |
| DENG Z J, SONG S Q, AI X R, YAO L. Principles and Techniques of Plant Seed Conservation and Test. Beijing: Science Press, 2019. ( in Chinese) | |
| [3] | FINKELSTEIN R, REEVES W, ARIIZUMI T, SREBER C . Molecular aspects of seed dormancy. Annual Review of Plant Biology, 2008,59:387-415. |
| [4] | SHU K, LIU X D, XIE Q, HE Z H . Two faces of one seed: Hormonal regulation of dormancy and germination. Molecular Plant, 2016,9:34-45. |
| [5] | 宋松泉 . 种子休眠//“10000个科学难题”农业科学编委会. 10000个科学难题. 北京: 科学出版社, 2011: 31-35. |
| SONG S Q. Seed dormancy//The Editorial Board of Agricultural Science for 10000 Selected Problems in Sciences, ed. 10000 Selected Problems in Sciences. Beijing: Science Press, 2011: 31-35. (in Chinese) | |
| [6] | GUBLER F, MILLER A A, JACOBSEN J V . Dormancy release, ABA and pre-harvest sprouting. Current Opinion in Plant Biology, 2005,8:183-187. |
| [7] | NONOGAKI H . Seed dormancy and germination-emerging mechanism and new hypotheses. Frontiers in Plant Science, 2014,e5:233. |
| [8] | CORBINEAU F, XIA Q, BAILLY C, EI-MAAROUF-BOUTEAU H . Ethylene, a key factor in the regulation of seed dormancy. Frontiers in Plant Science, 2014,5:539. |
| [9] | KUCERA B, COHN M A, LEUBNER-METZGER G . Plant hormone interactions during seed dormancy release and germination. Seed Science Research, 2005,15:281-307. |
| [10] | 徐恒恒, 黎妮, 刘树君, 王伟青, 王伟平, 张红, 程红焱, 宋松泉 . 种子萌发及其调控的研究进展. 作物学报, 2014,40:1141-1156. |
| XU H H, LI N, LIU S J, WANG W Q, WANG W P, ZHANG H, CHENG H Y, SONG S Q . Research progress in seed germination and its control. Acta Agronomica Sinica, 2014,40:1141-1156. (in Chinese) | |
| [11] | GRAEBER K, NAKABAYAYASHI K, MIATTON E, LEUBNER- METZGER G, SOPPE W J J . Molecular mechanisms of seed dormancy. Plant Cell and Environment, 2012,35:1769-1786. |
| [12] | CUTLER S R, RODRIGUEZ P L, FINKELSTEIN R R, ABRAMS S R . Abscisic acid: Emergence of a core signaling network. Annual Review of Plant Biology, 2010,61:651-679. |
| [13] | DEJONGHE W, OKAMOTO M, CUTLER S R . Small molecule probes of ABA biosynthesis and signaling. Plant Cell and Physiology, 2018,59:1490-1499. |
| [14] | VISHWAKARMA K, UPADHYAY N, KUMAR N, YADAV G, SINGH J, MISHRA R K, KUMAR V, VERMA R, UPADHYAY R G, PANDEY M, SHARMA S . Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Frontiers in Plant Science, 2017,8:161. |
| [15] | GIANINETTI A, VERNIERI P . On the role of abscisic acid in seed dormancy of red rice. Journal of Experimental Botany, 2007,58:3449-3462. |
| [16] | MILLAR A A, JACOBSEN J V, ROSS J J, HELLIWELL C A, POOLE A T, SCOFIELD G, REID J B, GUBLER F . Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8'-hydroxylase. The Plant Journal, 2006,45:942-954. |
| [17] | NAMBARA E, OKAMOTO M, TATEMATSU K, YANO R, SEO M, KAMIYA Y . Abscisic acid and the control of seed dormancy and germination. Seed Science Research, 2010,20:55-67. |
| [18] | NAMBARA E, MARION-POLL A . Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology, 2005,56:165-185. |
| [19] | NONOGAKI H . Seed germination and dormancy - The classic story, new puzzles, and evolution. Journal of Integrative Plant Biology, 2019,61:541-563. |
| [20] | HOLDSWORTH R, BENTSINK L, SOPPE W J J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist, 2008,179:33-54. |
| [21] | LINKIES A, LEUBNER-METZGER G . Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Reports, 2012,31:253-270. |
| [22] | FINKELSTEIN R . Abscisic acid synthesis and response. Arabidopsis Book, 2013,11:e0166. |
| [23] | NORTH H M, ALMEIDA A D, BOUTIN J-P, FREY A, TO A, BOTRAN L, SOTTA B, MARION-POLL A . The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. The Plant Journal, 2007,50:810-824. |
| [24] | TAN B C, JOSEPH L M, DENG W T, LIU L, LI Q B, CLINE K , McCARTY D R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. The Plant Journal, 2003,35:44-56. |
| [25] | CHENG W H, ENDO A, ZHOU L, PENNEY J, CHEN H C, ARROYO A, LEON P, NAMBARA E, ASAMI T, SEO M, KOSHIBA T, SHEEN J . A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. The Plant Cell, 2002,14:2723-2743. |
| [26] | GONZÁLEZ-GUZMÁ M, APOSTOLOVA N, BELLÉS J M, BARRERO J M, PIQUERAS P, PONCE M R, MICOL J L, SERRANO R, RODRÍGUEZ P . The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. The Plant Cell, 2002,14:1833-1846. |
| [27] | XIONG L, ISHITANI M, LEE H, ZHU J K . The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. The Plant Cell, 2001,13:2063-2083. |
| [28] | GAMBLE P E, MULLET J E . Inhibition of carotenoid accumulation and abscisic acid biosynthesis in fluridone-treated dark-grown barley. European Journal of Biochemistry, 1986,160:117-121. |
| [29] | CREELMAN R A, BELL E, MULLET J E . Involvement of a lipoxygenase-like enzyme in abscisic acid biosynthesis. Plant Physiology, 1992,99:1258-1260. |
| [30] | MÉRIGOUT P, KÉPÈS F, PERRET A M, SATIAT-JEUNEMAITRE B, MOREAU P . Effects of brefeldin A and nordihydroguaiaretic acid on endomembrane dynamics and lipid synthesis in plant cells. FEBS Letters, 2002,518:88-92. |
| [31] | HAN S Y, KITAHATA N, SEKIMATA K, SAITO T, KOBAYASHI M, NAKASHIMA K, YAMAGUCHI-SHINOZAKI K, SHINOZAKI K, YOSHIDA S, ASAMI T . A novel inhibitor of 9-cis-epoxycarotenoid dioxygenase in abscisic acid biosynthesis in higher plants. Plant Physiology, 2004,135:1574-1582. |
| [32] | KITAHATA N, HAN S Y, NOJI N, SAITO T, KOBAYASHI M, NAKANO T, KUCHITSU K, SHINOZAKI K, YOSHIDA S, MATSUMOTO S . A 9-cis-epoxycarotenoid dioxygenase inhibitor for use in the elucidation of abscisic acid action mechanisms. Bioorganic and Medicinal Chemistry, 2006,14:5555-5561. |
| [33] | BOYD J, GAI Y, NELSON K M, LUKIWSKI E, TALBOT J, LOEWEN M K, OWEN S, ZAHARIA L I, CUTLER A J, ABRAMS S R, LOEWEN M C . Sesquiterpene-like inhibitors of a 9-cis- epoxycarotenoid dioxygenase regulating abscisic acid biosynthesis in higher plants. Bioorganic and Medicinal Chemistry, 2009,17:2902-2912. |
| [34] | KUSHIRO T, OKAMOTO M, NAKABAYASHI K, YAMAGISHI K, KITAMURA S, ASAMI T, HIRAI N, KOSHIBA T, KAMIYA Y, NAMBARA E . The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: Key enzymes in ABA catabolism. The EMBO Journal, 2004,23:1647-1656. |
| [35] | SAITO S, HIRAI N, MATSUMOTO C, OHIGASHI H, OHTA D, SAKATA K, MIZUTANI M . Arabidopsis CYP707As encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiology, 2004,134:1439-1449. |
| [36] | HANADA K, HASE T, TOYODA T, SHIONZAKI K, OKAMOTO M . Origin and evolution of genes related to ABA metabolism and its signaling pathways. Journal of Plant Research, 2011,124:455-465. |
| [37] | OKAMOTO M, KUWAHARA A, SEO M, KUSHIRO T, ASAMI T, HIRAI N, KAMIYA Y, KOSHIBA T, NAMBARA E . CYP707A1 and CYP707A2, which encode ABA 8'-hydroxylases, are indispensable for a proper control of seed dormancy and germination in Arabidopsis. Plant Physiology, 2006,141:97-107. |
| [38] | OKAMOTO M, TANAKA Y, ABRAMS S R, KAMIYA Y, SEKI M, NAMBARA E . High humidity induces abscisic acid 8'-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiology, 2009,149:825-834. |
| [39] | KEPKA M, BENSON C L, GONUGUNTA V K, NELSON K M, CHRISTMANN A, GRILL E, ABRAMA S R . Action of natural abscisic acid precursors and catabolites on abscisic acid receptor complexes. Plant Physiology, 2011,157:2108-2119. |
| [40] | WENG J K, YE M, LI B, NOEL J P . Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell, 2016,166:881-893. |
| [41] | KITAHATA N, SAITO S, MIYAZAWA Y, UMEZAWA T, SHIMADA Y, MIN Y K, MIZUTANI M, HIRAI N, SHINOZAKI K, YOSHIDA S . Chemical regulation of abscisic acid catabolism in plants by cytochrome P450 inhibitors. Bioorganic and Medicinal Chemistry, 2005,13:4491-4498. |
| [42] | SAITO S, OKAMOTO M, SHINODA S, KUSHIRO T, KOSHIBA T, KAMIYA Y, HIRAI N, TODOROKI Y, SAKATA K, NAMBARA E, MIZUTANI M . A plant growth retardant, uniconazole, is a potent inhibitor of ABA catabolism in Arabidopsis. Bioscience Biotechnology and Biochemistry, 2006,70:1731-1739. |
| [43] | RADEMACHER W . Growth retardants: Effects on gibberellin biosynthesis and other metabolic pathways. Annual Review of Plant Physiology and Plant Molecular Biology, 2000,51:501-531. |
| [44] | TAKEUCHI J, OKAMOTO M, MEGA R, KANNO Y, OHNISHI T, SEO M, TODOROKI Y . Abscinazole-E3M, a practical inhibitor of abscisic acid 8'-hydroxylase for improving drought tolerance. Scientific Reports, 2016,6:37060. |
| [45] | BENSON C L, KEPKA M, WUNSCHEL C, RAJAGOPALAN N, NELSON K M, CHRISTMANN A, ABRAMS S R, GRILL E, LOEWEN M . Abscisic acid analogs as chemical probes for dissection of abscisic acid responses in Arabidopsis thaliana. Phytochemistry, 2015,113:96-107. |
| [46] | ARAKI Y, MIYAWAKI A, MIYASHITA T, MIZUTANI M, HIRAI N, TODOROKI Y . A new non-azole inhibitor of ABA 8'-hydroxylase: Effect of the hydroxyl group substituted for geminal methyl groups in the six-membered ring. Bioorganic and Medicinal Chemistry Letters, 2006,16:3302-3305. |
| [47] | MA Y, SZOSTKIEWICZ I, KORTE A, MOES D, YANG Y, CHRISTMANN A, GRILL E . Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 2009,324:1064-1068. |
| [48] | NISHIMURA N, TSUCHIYA W, MORESCO J J, HAYASHI Y, SATOH K, KAIWA N, IRISA T, KINOSHITA T, SCHROEDER J I, YATES J R, HIRAYAMA T, YAMAZAKI T . Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nature Communication, 2018,9:2132. |
| [49] | PARK S Y, FUNG P, NISHIMURA N, JENSEN D R, FUJII H, ZHAO Y, LUMBA S, SANTIAGO J, RODRIGUES A, CHOW T F, ALFRED S E, BONETTA D, FINKELSTEIN R, PROVART N J, DESVEAUX D, RODRIGUEZ P L, McCOURT P, ZHU J K, SCHROEDER J I, VOLKMAN B F, CUTLER S R . Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 2009,324:1068-1071. |
| [50] | MELCHER K, NG L M, ZHOU X E, SOON F-F, XU Y, SUINO-POWELL K M, PARK S Y, WEINER J J, FUJII H, CHINNUSAMY V, KOVACH A, LI J, WANG Y, LI J, PERTERSON F C, JENSEN D R, YONG E L, VOLKMAN B F, CUTLER S R, ZHU J K, XU H E. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature, 2009,462:602-608. |
| [51] | SANTIAGO J, DUPEUX F, ROUND A, ANTONI R, PARK S Y, JAMIN M, CUTLER S R, RODRIGUEZ P L, MÁRQUEZ J A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature, 2009,462:665-668. |
| [52] | YIN P, FAN H, HAO Q, YUAN X, WU D, PANG Y, YAN C, LI W, WANG J, YAN N . Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nature Structural and Molecular Biology, 2009,16:1230-1236. |
| [53] | FUJII H, ZHU J K . Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences of the United States of America, 2009,106:8380-8385. |
| [54] | SOON F F, NG L M, ZHOU X E, WEST G M, KOVACH A, TAN M H E, SUINO-POWELL K M, HE Y, XU Y, CHALMERS M J, BRUNZELLE J S, ZHANG H, YANG H, JIANG H, LI J, YONG E L, CUTLER S, ZHU J K, GRIFFIN P R, MELCHER K, XU H E. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science, 2012,335:85-88. |
| [55] | NAKAMURA S, LYNCH T J, FINKELSTEIN R R . Physical interactions between ABA response loci of Arabidopsis. The Plant Journal, 2001,26:627-635. |
| [56] | HAUSER F, WAADT R, SCHROEDER J I . Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology, 2011,21:R346-R355. |
| [57] | BOWMAN J L, KOHCHI T, YAMATO K T, JENKINS J, SHU S, ISHIZAKI K, YAMAOKA S, NISHIHAMA R, NAKAMURA Y, BERGER F, ADAM C, AKI S S, ALTHOFF F, ARAKI T, ARTEAGA-VAZQUEZ M A, BALASUBRMANIAN S, BARRY K, BAYER D, SCHMUTZ J. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell, 2017,171:287-304. |
| [58] | TISCHER S V, WUNSCHEL C, PAPACEK M, KLEIGREWE K, HOFMANN T, CHRISTMANN A, GRILL E . Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 2017,114:10280-10285. |
| [59] | ANTONI R, GONZALEZ-GUZMAN M, RODRIGUEZ L, RODRIGUES A, PIZZIO G A, RODRIGUEZ P L . Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiology, 2012,158:970-980. |
| [60] | NISHIMURA N, YOSHIDA T, KITAHATA N, ASAMI T, SHINOZAKI K, HIRAYAMA T . ABA-HYPERSENSITIVE GERMINATION1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. The Plant Journal, 2007,50:935-949. |
| [61] | FUJITA Y, NAKASHIMA K, YOSHIDA T, KATAGIRI T, KIDOKORO S, KANAMORI N, UMEZAWA T, FUJITA M, MARUYAMA K, ISHIYAMA K, KOBAYASHI M, NAKASONE S, YAMADA K, ITO T, SHINOZAKI K . Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant and Cell Physiology, 2009,50:2123-2132. |
| [62] | NAKASHIMA K, FUJITA Y, KANAMORI N, KATAGIRI T, UMEZAWA T, KIDOKORO S, MARUYAMA K, YOSHIDA T, ISHIYAMA K, KOBAYASHI M, SHINOZAKI K, YAMAGUCHI- SHINOZAKI K . Three Arabidopsis SnRK2 protein kinases, SRK2D/ SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant and Cell Physiology, 2009,50:1345-1363. |
| [63] | UMEZAWA T, SUGIYAMA N, MIZOGUCHI M, HAYASHI S, MYOUGA F, YAMAGUCHI-SHINOZAKI K, ISHIHAMA Y, HIRAYAMA T, SHINOZAKI K . Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences of the United states of America, 2009,106:17588-17593. |
| [64] | KLINE K G, BARRETT-WILT G A, SUSSMAN M R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proceedings of the National Academy of Sciences of the United States of America, 2010,107:15986-15991. |
| [65] | UMEZAWA T, SUGIYAMA N, TAKAHASHI F, ANDERSON J C, ISHIHAMA Y, PECK S C, SHINOZAKI K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Science Signaling, 2013, 6:1rs8. |
| [66] | WANG P, XUE L, BATELLI G, LEE S, HOU Y J, VAN OOSTEN M J, ZHANG H, TAO W A, ZHU J K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proceedings of the National Academy of Sciences of the United states of America, 2013,110:11205-11210. |
| [67] | DING S, ZHANG B, QIN F . Arabidopsis RZFP34/CHYR1, an ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. The Plant Cell, 2015,27:3228-3244. |
| [68] | BELDA-PALAZON B, RODRIGUEZ L, FERNANDEZ M A, CASTILLO M C, ANDERSON E A, GAO C, GONZALEZ- GUZMAN M, PEIRATS-LLOBET M, ZHAO Q, DE WINNE N, GEVEERT K, DE JAEGER G, JIANG L, LEÒN J, MULLEN R T, RODRIGUEZ P L. FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. The Plant Cell, 2016,28:2291-2311. |
| [69] | WU Q, ZHANG X, PEIRATS-LLOBET M, BELDA-PALAZON B, WANG X, CUI S, YU X, RODRIGUEZ P L, AN C . Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. The Plant Cell, 2016,28:2178-2196. |
| [70] | YU F, WU Y, XIE Q . Ubiquitin-proteasome system in ABA signaling: From perception to action. Molecular Plant, 2016,9:21-33. |
| [71] | ZHAO J, ZHAO L, ZHANG M, ZAFAR S, FANG J, LI M, ZHANG W, LI X . Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation. International Journal of Molecular Science, 2017,18:1841. |
| [72] | KARSSEN C M, BRINKHORST-VAN DER SWAN D L C, BREEKLAND A E, KOORNNEEF M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta, 1983,157:158-165. |
| [73] | FREY A, GODIN B, BONNET M, SOTTA B, MARION-POLL A . Maternal synthesis of abscisic acid controls seed development and yield in Nicotiana plumbaginifolia. Planta, 2004,218:958-964. |
| [74] | KOORNNEEF M, HANHART C J, HILHORST H W M, KARSSEN C M. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiology, 1989,90:463-469. |
| [75] | CADMAN C S, TOOROP P E, HILHORST H W M, FINCH- SAVAGE W E. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal, 2006,46:805-822. |
| [76] | LEFEBVRE V, NORTH H, FREY A, SOTTA B, SEO M, OKAMOTO M, NAMBARA E, MARION-POLL A . Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesised in the endosperm is involved in the induction of seed dormancy. The Plant Journal, 2006,45:309-319. |
| [77] | NAKABAYASHI K, OKAMOTO M, KOSHIBA T, KAMIYA Y, NAMBARA E . Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. The Plant Journal, 2005,41:697-709. |
| [78] | FINCH-SAVAGE W E, LEUBNER-METZGER G . Seed dormancy and the control of germination. New Phytologist, 2006,171:501-523. |
| [79] | GUBLER F, HUGHES T, WATERHOUSE P, JACOBSEN J . Regulation of dormancy in barley by blue light and after-ripening: Effects on abscisic acid and gibberellin metabolism. Plant Physiology, 2008,147:886-896. |
| [80] | JACOBSEN J V, BARRERO J M, HUGHES T, JULKOWSKA M, TAYLOR J M, XU Q, GUBLER F . Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat (Triticum aestivum L.) grain. Planta, 2013,238:121-138. |
| [81] | LIU A, GAO F, KANNO Y, JORDAN M C, KAMIYA Y, SEO M, AYELE B T . Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE, 2013,8:e56570. |
| [82] | BARRERO J M, TALBOT M J, WHITE R G, JACOBSEN J V, GUBLER F . Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiology, 2009,150:1006-1021. |
| [83] | LIU Y, SHI L, YE N, LIU R, JIA W, ZHANG J . Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytologist, 2009,183:1030-1042. |
| [84] | PRESTON J, TATEMATSU K, KANNO Y, HOBO T, KIMURA M, JIKUMARU Y, YANO R, KAMIYA Y, NAMBARA E . Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: A comparative study on dormant and non-dormant accessions. Plant and Cell Physiology, 2009,50:1786-1800. |
| [85] | MATAKIADIS T, ALBORESI A, JIKUMARU Y, TATEMATSU K, PICHON O, RENOU J P, SOTTA B, KAMIYA Y, NAMBARA E, TROUNG H N . The Arabidopsis abscisic acid catabolism gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiology, 2009,149:949-960. |
| [86] | TOH S, IMAMURA A, WATANABE A, NAKABAYASHI K, OKAMOTO M, JIKUMARU Y, HANADA A, ASO Y, ISHIYAMA K, TAMURA N, IUCHI S, KOBAYASHI M, YAMAGUCHI S, KAMIYA Y, NAMBARA E, KAWAKAMI N . High temperature- induced ABA biosynthesis and its role in the inhibition of GA action in Arabidopsis seeds. Plant Physiology, 2008,146:1368-1385. |
| [87] | BENECH-ARNOLD R L, GUALANO N, LEYMARIE J, CÔME D, CORBINEAU F . Hypoxia interferes with ABA metabolism and increases ABA sensitivity in embryos of dormant barley grains. Journal of Experimental Botany, 2006,57:1423-1430. |
| [88] | TOOROP P E, BEWLEY J D, HILHORST H W M. Endo-β-isoforms are present in the endosperm and embryo of tomato seeds, but are not essentially linked to germination. Planta, 1996,200:153-158. |
| [89] | MÜLLER K, TINTELNOT S, LEUBNER-METZGER G . Endosperm- limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum(cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant and Cell Physiology, 2006,47:864-877. |
| [90] | MÜLLER K, CARSTENS A C, LINKIES A, TORRES M A, LEUBNER-METZGER G . The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytologist, 2009,184:885-897. |
| [91] | NONOGAKI H . Seed biology updates-highlights and new discoveries in seed dormancy and germination research. Frontiers in Plant Science, 2017,8:524. |
| [92] | NÉE G, KRAMER K, NAKABAYASHI K, YUAN B, XIANG Y, MIATTON E, FINKEMEIER I, SOPPE W J . DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nature Communication, 2017,8:72. |
| [93] | ALONSO-BLANCO C, BENTSINK L, HANHART C J, VRIES H B D, KOORNNEEF M . Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics, 2003,164:711-729. |
| [94] | BENTSINK L, HANSON J, HANHART C J, BLANKESTIJN-DE VRIES H, COLTRANE C, KEIZER P, EL-LITHY M, ALONSO-BLANCO C, DE ANDRÉS M T, REYMOND M, VAN EEUWIJK F, SMEEKENS S, KOORNNEEF M. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proceedings of the National Academy of Sciences of the United States of America , 2010,107:4264-4269. |
| [95] | BENTSINK L, JOWETT J, HANHART C J, KOORNNEEF M . Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 2006,103:17042-17047. |
| [96] | NAKABAYASHI K, BARTSCH M, XIANG Y, MIATTON E, PELLENGAHR S, YANO R, SEO M, SOPPE W J J. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. The Plant Cell, 2012,24:2826-2838. |
| [97] | NAKABAYASHI K, BARTSCH M, DING J, SOPPE W J . Seed dormancy in Arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genetics, 2015,11:e1005737. |
| [98] | CYREK M, FEDAK H, CIESIELSKI A, GUO Y W, SLIWA A, BRZEZNIAK L, KRZYCZMONIK K, PIETRAS Z, KACZANOWSKI S, LIU F, SWIEZEWSKI S . Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiology, 2016,170:947-955. |
| [99] | DOLATA J, GUO Y, KOŁOWERZO A, SMOLIŃSKI D, BRZYŻEK G, JARMOŁOWSKI A, ŚWIEŻEWSKI S. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. The EMBO Journal, 2015,34:544-558. |
| [100] | LIU Y, KOORNNEEF M, SOPPE W J . The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. The Plant Cell, 2007,19:433-444. |
| [101] | LIU Y, GEYER R, VAN ZANTEN M, CARLES A, LI Y, HÖROLD A, VAN NOCKER S, SOPPE W J J. Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE, 2011,6:e22241. |
| [102] | MORTENSEN S A, GRASSER K D . The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Letters, 2014,588:47-51. |
| [103] | DI GIAMMARTINO D C, NISHIDA K, MANLEY J L . Mechanisms and consequences of alternative polyadenylation. Molecular Cell, 2011,43:853-866. |
| [104] | FEDAK H, PALUSINSKA M, KRZYCZMONIK K, BRZEZNIAK L, YATUSEVICH R, PIETRAS Z, KACZANOWSKI S, SWIEZEWSKI S . Control of seed dormancy in Arabidopsis by a cis-acting non- coding antisense transcript. Proceedings of the National Academy of Sciences of the United States of America, 2016,113:E7846-E7855. |
| [105] | ARCHACKI R, YATUSEVICH R, BUSZEWICZ D, KRZYCZMONIK K, PATRYN J, IWANICKA-NOWICKA R, BIECEK P, WILCZYNSKI B, KOBLOWSKA M, JERZMANOWSKI A, SWIEZEWSKI S . Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Research, 2016,45:3116-3129. |
| [106] | LIN S, ZHANG L, LUO W, ZHANG X . Characteristics of antisense transcript promoters and the regulation of their activity. International Journal of Molecular Sciences, 2015,17:1-17. |
| [107] | KORNIENKO A E, GUENZL P M, BARLOW D P, PAULER F M . Gene regulation by the act of long non-coding RNA transcription. BMC Biology, 2013,11:59. |
| [108] | PELECHANO V, STEINMETZ L M . Non-coding RNA gene regulation by antisense transcription. Nature Review of Genetics, 2013,14:880-893. |
| [109] | QUINN J J, CHANG H Y . Unique features of long non-coding RNA biogenesis and function. Nature Review Genetics, 2016,17:47-62. |
| [110] | SHEARWIN K E, CALLEN B P, EGAN J B . Transcriptional interference - a crash course. Trends in Genetics, 2005,21:339-345. |
| [111] | HONGAY C F, GRISAFI P L, GALITSKI T, FINK G R . Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell, 2006,127:735-745. |
| [112] | NÉE G, XIANG Y, SOPPE W J . The release of dormancy, a wake-up call for seeds to germinate. Current Opinion in Plant Biology, 2016,35:8-14. |
| [113] | YOSHIDA T, NISHIMURA N, KITAHATA N, KUROMORI T, ITO T, ASAMI T, SHINOZAKI K, HIRAYAMA T . ABA-HYPERSENSITIVE GERMINATION3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiology, 2005,140:115-126. |
| [114] | LI T, BONKOVSKY H L, GUO J . Structural analysis of heme proteins: Implications for design and prediction. BMC Structural Biology, 2011,11:13. |
| [115] | ALBERTOS P, ROMERO-PUERTAS M C, TATEMATSU K, MATEOS I, SANCHEZ-VICENTE I, NAMBARA E, LORENZO O, SÁNCHEZ-VICENTE I, NAMBARA E, LORENZO O. S- Nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nature Communication, 2015,6:1-10. |
| [116] | OHKUMA K, LYON J L, ADDICOTT F T, SMITH O E . Abscisin II, an abscission-accelerating substance from young cotton fruit. Science, 1963,142:1592-1593. |
| [117] | WANG Y G, FU F L, YU H Q, HU T, ZHANG Y Y, TAO Y, ZHU J K, ZHAO Y, LI W C . Interaction network of core ABA signaling components in maize. Plant Molecular Biology, 2018,96:245-263. |
| [118] | LIU S J, SONG S H, WANG W Q, SONG S Q . De novo assembly and characterization of germinating lettuce seed transcriptome using Illumina paired-end sequencing. Plant Physiology and Biochemistry, 2015,96:154-162. |
| [119] | WANG W Q, SONG B Y, DENG Z J, WANG Y, LIU S J, MØLLER I M, SONG S Q . Proteomic analysis of Lactuca sativa seed germination and thermoinhibition by sampling of individual seeds at germination and removal of storage proteins by PEG fractionation. Plant Physiology, 2015,167:1332-1350. |
| [120] | XU H H, LIU S J, SONG S H, WANG W Q, MØLLER I X, SONG S Q . Proteome changes associated with dormancy release of Dongxiang wild rice seeds. Journal of Plant Physiology, 2016,206:68-86. |
| [1] | 唐玉林, 张博, 任曼, 张瑞雪, 秦俊杰, 朱浩, 郭延生. UPLC-MS/MS代谢组学评价归芪益母口服液对产后奶牛瘤胃的调节作用[J]. 中国农业科学, 2023, 56(2): 368-378. |
| [2] | 林馨颖,王鹏杰,杨如兴,郑玉成,陈潇敏,张磊,邵淑贤,叶乃兴. 高茶氨酸茶树新品系‘福黄1号’黄化变异机理[J]. 中国农业科学, 2022, 55(9): 1831-1845. |
| [3] | 李青林,张文涛,徐慧,孙京京. 低磷胁迫下黄瓜木质部与韧皮部汁液的代谢物变化[J]. 中国农业科学, 2022, 55(8): 1617-1629. |
| [4] | 谢意通,张飞,石洁,冯莉,姜丽. 外源蔗糖对紫背天葵采后品质及叶绿体的影响[J]. 中国农业科学, 2022, 55(8): 1642-1656. |
| [5] | 余琦隆,韩莹琰,郝敬虹,秦晓晓,刘超杰,范双喜. 外源亚精胺对高温胁迫下生菜氮代谢的影响[J]. 中国农业科学, 2022, 55(7): 1399-1410. |
| [6] | 吕馨宁,王玥,贾润普,王胜男,姚玉新. 不同温度下褪黑素处理对‘阳光玫瑰'葡萄采后品质的影响[J]. 中国农业科学, 2022, 55(7): 1411-1422. |
| [7] | 宋松泉,刘军,唐翠芳,程红焱,王伟青,张琪,张文虎,高家东. 种子耐脱水性的生理及分子机制研究进展[J]. 中国农业科学, 2022, 55(6): 1047-1063. |
| [8] | 闫乐乐,卜璐璐,牛良,曾文芳,鲁振华,崔国朝,苗玉乐,潘磊,王志强. 广泛靶向代谢组学解析桃蚜危害对桃树次生代谢产物的影响[J]. 中国农业科学, 2022, 55(6): 1149-1158. |
| [9] | 彭佳堃, 戴伟东, 颜涌泉, 张悦, 陈丹, 董明花, 吕美玲, 林智. 基于代谢组学的‘永春佛手’乌龙茶化学成分解析[J]. 中国农业科学, 2022, 55(4): 769-784. |
| [10] | 宋江涛,谌丹丹,公旭晨,商祥明,李春龙,蔡永喜,岳建平,王帅玲,张卜芬,谢宗周,刘继红. 人工疏果对‘爱媛28’橘橙果实糖酸含量及代谢基因表达的影响[J]. 中国农业科学, 2022, 55(23): 4688-4701. |
| [11] | 王娟,陈皓宁,石大川,于天一,闫彩霞,孙全喜,苑翠玲,赵小波,牟艺菲,王奇,李春娟,单世华. 花生高亲和硝酸盐转运蛋白基因AhNRT2.7a响应低氮胁迫的功能研究[J]. 中国农业科学, 2022, 55(22): 4356-4372. |
| [12] | 尤佳玲,李有梅,孙孟豪,谢兆森. ‘黑比诺’葡萄不同叶龄叶片叶绿体内淀粉积累及其相关基因表达差异分析[J]. 中国农业科学, 2022, 55(21): 4265-4278. |
| [13] | 孙保娟,汪瑞,孙光闻,王益奎,李涛,宫超,衡周,游倩,李植良. 转录组及代谢组联合解析茄子果色上位遗传效应[J]. 中国农业科学, 2022, 55(20): 3997-4010. |
| [14] | 李刚,白阳,贾子颖,马正阳,张祥池,李春艳,李诚. 两种磷素水平下小麦苗期对干旱胁迫的离子组和代谢组响应[J]. 中国农业科学, 2022, 55(2): 280-294. |
| [15] | 耿文杰,李宾,任佰朝,赵斌,刘鹏,张吉旺. 种植密度和喷施乙烯利对夏玉米木质素代谢和抗倒伏性能的调控[J]. 中国农业科学, 2022, 55(2): 307-319. |
|
||