













中国农业科学 ›› 2024, Vol. 57 ›› Issue (8): 1417-1429.doi: 10.3864/j.issn.0578-1752.2024.08.001
收稿日期:2023-10-23
接受日期:2023-11-29
出版日期:2024-04-16
发布日期:2024-04-24
通信作者:
联系方式:
许娜,E-mail:xuna1109@163.com。
基金资助:
XU Na( ), TANG Ying, XU ZhengJin, SUN Jian(
), TANG Ying, XU ZhengJin, SUN Jian( ), XU Quan(
), XU Quan( )
)
Received:2023-10-23
Accepted:2023-11-29
Published:2024-04-16
Online:2024-04-24
摘要:
【目的】籼粳亚种间杂交F1的育性问题严重阻碍了亚种间杂种优势的利用,探究籼粳杂种不育的遗传机理,挖掘新的籼粳杂种不育调控基因,为促进籼粳杂种结实率遗传改良提供理论依据。【方法】粳稻品种Sasanishiki和籼稻品种Habataki杂交后,采用单粒传法自交10代,获得包含95个株系的稳定遗传重组自交系(RIL),基于Illumina平台,对双亲和RIL进行高通量测序,在全基因组水平分析RIL中Habataki血缘的分布与比例,进而鉴定偏分离区域作为潜在的籼粳杂种不育位点。同时,剖析“全球3000份水稻核心种质资源重测序计划”中典型籼粳稻基因组变异数据,对群体水平进一步验证并趋近目标育性基因区域。最终通过序列比对锁定籼粳杂种不育候选基因。应用CRISPR基因编辑技术进行定点基因敲除,对目标基因进行功能验证。【结果】Habataki和Sasanishiki的杂交F1在穗数、每穗粒数和千粒重上体现出明显的杂种优势,但其结实率显著降低,I2-KI镜检发现F1花粉育性显著降低。RIL的全基因组高通量测序在第1、3、5、6、7和12染色体上检测到明显的偏分离,即该区域的基因型趋向于籼稻Habataki。通过序列比对,进一步确定已知育性基因Sc、S5和HSA1为第3、6和12染色体上偏分离区域的目标基因。应用CRISPR基因编辑技术敲除Habataki中Sc-Haba-3的多拷贝,成功改善了其与Sasanishiki杂交F1的花粉育性和结实率,说明该基因可独立行使功能。同时,在第1染色体的偏分离区域发现Habataki和Sasanishiki之间存在复杂的结构性变异,Sasanishiki基因组中一段24.7 kb包含4个预测基因的片段在Habataki中被一段64.8 kb包含10个预测基因的片段取代,此结构性变异可能参与籼粳杂种育性的调控。【结论】检测到多个籼粳杂种不育相关位点,应用CRISPR基因编辑技术敲除Habataki中Sc的多拷贝成功改善其杂种F1育性,确定其为水稻亚种间育性改良的目标基因。同时锁定了第1染色体Sd区域的目标基因。
许娜, 唐颖, 徐正进, 孙健, 徐铨. 籼粳杂种不育的遗传分析和候选基因鉴定[J]. 中国农业科学, 2024, 57(8): 1417-1429.
XU Na, TANG Ying, XU ZhengJin, SUN Jian, XU Quan. Genetic Analysis and Candidate Gene Identification on Fertility and Inheritance of Hybrid Sterility of XI and GJ Cross[J]. Scientia Agricultura Sinica, 2024, 57(8): 1417-1429.
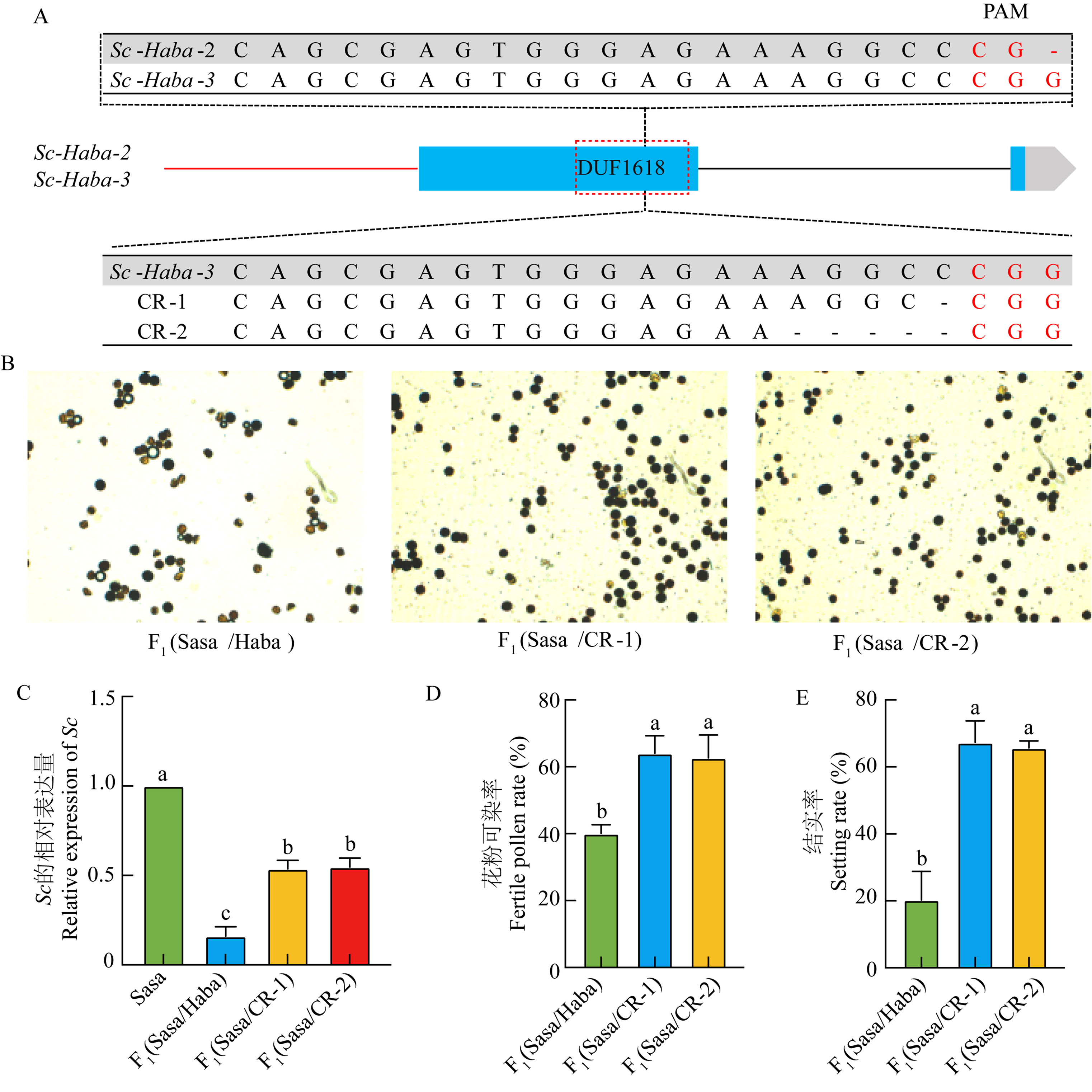
图6
Haba中Sc-Haba-3的CRISPR基因编辑植株的突变序列和表型分析 A:Sc-Haba-2与Sc-Haba-3靶点PAM序列的差异和基因编辑植株的突变序列;B:F1(Sasa/Haba)、F1(Sasa/CR-1)和F1(Sasa/CR-2)植株的花粉I2-KI镜检;C:Sasa、F1(Sasa/Haba)、F1(Sasa/CR-1)和F1(Sasa/CR-2)中Sc-Sasa的表达量;D:F1(Sasa/Haba)、F1(Sasa/CR-1)和F1(Sasa/CR-2)的花粉可染率;E:F1(Sasa/Haba)、F1(Sasa/CR-1)和F1(Sasa/CR-2)的结实率"
| [1] |
doi: 10.1534/genetics.104.035642 |
| [2] |
doi: 10.1038/ng.695 pmid: 20972439 |
| [3] |
doi: 10.1038/ng.1018 |
| [4] |
doi: 10.1016/j.cell.2021.01.013 pmid: 33539781 |
| [5] |
doi: 10.1038/s41586-018-0063-9 |
| [6] |
doi: 10.1038/s41588-020-00769-9 pmid: 33526925 |
| [7] |
doi: 10.1038/s41586-020-03091-w |
| [8] |
|
| [9] |
袁隆平. 杂交水稻的育种战略设想. 杂交水稻, 1987(1): 1-3.
|
|
|
|
| [10] |
doi: 10.1093/aob/mcm121 |
| [11] |
doi: 10.1093/genetics/77.3.521 pmid: 17248657 |
| [12] |
doi: 10.1093/genetics/125.1.183 pmid: 2341030 |
| [13] |
|
| [14] |
doi: 10.1038/s41467-017-01400-y pmid: 29101356 |
| [15] |
doi: 10.1126/science.1223702 pmid: 22984070 |
| [16] |
doi: 10.1016/j.molp.2015.09.014 |
| [17] |
|
| [18] |
doi: 10.1016/j.cell.2023.06.023 |
| [19] |
doi: 10.1186/s12915-018-0572-x |
| [20] |
doi: 10.1007/s00122-008-0723-5 |
| [21] |
张桂权, 卢永根, 张华, 杨进昌, 刘桂富. 栽培稻(Oryza sativa)杂种不育性的遗传研究: Ⅳ.F1花粉不育性的基因型. 遗传学报, 1994, 21(1): 34-41.
|
|
|
|
| [22] |
doi: 10.1007/s11434-006-0675-6 |
| [23] |
doi: 10.1016/S2095-3119(19)62843-1 |
| [24] |
doi: 10.1038/s41467-023-40189-x |
| [25] |
doi: 10.1016/j.molp.2023.02.009 |
| [26] |
doi: 10.1016/j.cell.2021.04.046 pmid: 34051138 |
| [27] |
doi: S1360-1385(19)30015-9 pmid: 30745056 |
| [28] |
doi: 10.1038/s41422-022-00685-z pmid: 35821092 |
| [29] |
doi: 10.1111/pbi.v21.1 |
| [30] |
doi: 10.1038/ng.3346 |
| [31] |
doi: 10.1371/journal.pgen.1006386 |
| [32] |
doi: 10.1104/pp.104.045039 |
| [33] |
吴疆. 水稻Sc座位蛋白复合体参与籼粳杂种雄性不育的研究[D]. 广州: 华南农业大学, 2017.
|
|
|
|
| [34] |
袁隆平. 超级杂交水稻育种研究新进展. 中国农村科技, 2010, Z1: 24-25.
|
|
|
|
| [35] |
doi: 10.1111/jipb.v59.9 |
| [1] | 陈兵先, 张琪, 戴彰言, 周旭, 刘军. 水杨酸引发提高低温下水稻种子萌发活力的生理与分子效应[J]. 中国农业科学, 2024, 57(7): 1220-1236. |
| [2] | 宋雅荣, 常单娜, 周国朋, 高嵩涓, 段廷玉, 曹卫东. 解磷细菌活化水稻土中低品位磷矿粉的效果与机制[J]. 中国农业科学, 2024, 57(6): 1102-1116. |
| [3] | 张必东, 林泓, 朱思颖, 李忠成, 庄慧, 李云峰. 水稻颖壳异常突变体ah1的鉴定与候选基因分析[J]. 中国农业科学, 2024, 57(3): 429-441. |
| [4] | 刘苡含, 牟青山, 贺响, 陈敏, 胡晋, 关亚静. OsFWL3调控水稻金属离子转运和积累的研究[J]. 中国农业科学, 2024, 57(21): 4161-4174. |
| [5] | 蔡玉彪, 张坤杰, 王雅宣, 赖凤香, 何佳春, 万品俊, 傅强. 水稻品种对褐飞虱趋向二化螟危害株习性的影响[J]. 中国农业科学, 2024, 57(20): 3998-4006. |
| [6] | 郭乃辉, 张文忠, 圣忠华, 胡培松. 利用CRISPR/Cas9技术编辑MODD增强水稻休眠性[J]. 中国农业科学, 2024, 57(2): 227-235. |
| [7] | 张亚玲, 付忠举, 李雪, 孙宇佳, 赵羽涵, 顾欣怡, 王艳霞, 靳学慧, 吴伟怀, 华丽霞. 黑龙江、四川和海南省稻作区水稻穗腐病病原比较分析[J]. 中国农业科学, 2024, 57(2): 278-294. |
| [8] | 周信雁, 陈思宇, 魏宇飞, 朱瑜, 冯君茜, 丁点草, 陆贵锋, 杨尚东. 红皮白肉与红皮红肉火龙果植株茎部内生微生物群落结构特征[J]. 中国农业科学, 2024, 57(2): 416-428. |
| [9] | 武玉花, 翟杉杉, 普豪祯, 高鸿飞, 张华, 李俊, 李允静, 肖芳, 吴刚, 徐利群. 农业用植物基因编辑产品检测方法研究进展[J]. 中国农业科学, 2024, 57(17): 3318-3334. |
| [10] | 翁文安, 邢志鹏, 胡群, 魏海燕, 史扬杰, 奚小波, 李秀丽, 刘桂云, 陈娟, 袁风萍, 孟轶, 廖萍, 高辉, 张洪程. 无人化旱直播模式对水稻产量、品质和经济效益的影响[J]. 中国农业科学, 2024, 57(17): 3350-3365. |
| [11] | 苏素苗, 康天恺, 邹家龙, 汪本福, 张洋洋, 廖世鹏, 李小坤. 不同水稻品种和施锌方式对水稻产量及籽粒锌有效性的影响[J]. 中国农业科学, 2024, 57(15): 3023-3034. |
| [12] | 鄢柳慧, 钟琦, 马增凤, 韦敏益, 刘驰, 秦媛媛, 周小龙, 黄大辉, 卢颖萍, 秦钢, 张月雄. 普通野生稻早抽穗基因OsEHD8的鉴定与进化分析[J]. 中国农业科学, 2024, 57(14): 2703-2716. |
| [13] | 张建桃, 黄路生, 刘广彬, 兰玉彬, 文晟. 农药液滴在水稻叶面的动态润湿铺展行为[J]. 中国农业科学, 2024, 57(13): 2583-2598. |
| [14] | 舒小伟, 王树深, 伏桐, 王子涵, 丁周宇, 杨英, 赵士茹, 周娟, 黄建晔, 姚友礼, 王余龙, 董桂春. 小麦秸秆还田后不同水稻品种对简化穗肥施用的响应及其成因[J]. 中国农业科学, 2024, 57(10): 1961-1978. |
| [15] | 房康睿, 龙世平, 彭斯文, 陈山, 廖育林, 徐新朋, 赵士诚, 仇少君, 何萍, 周卫. 洞庭湖区典型稻田玉米水稻轮作下土壤—作物系统对施氮措施的响应[J]. 中国农业科学, 2024, 57(10): 1979-1994. |
|
||