













中国农业科学 ›› 2022, Vol. 55 ›› Issue (7): 1458-1468.doi: 10.3864/j.issn.0578-1752.2022.07.016
耿仁浩1,2( ),刘博1,王芳1,罗玉峰1,曲鸿飞1,范学政1,秦玉明1,丁家波1,许冠龙1,沈青春1(
),刘博1,王芳1,罗玉峰1,曲鸿飞1,范学政1,秦玉明1,丁家波1,许冠龙1,沈青春1( ),秦爱建2(
),秦爱建2( )
)
收稿日期:2021-02-08
接受日期:2021-06-21
出版日期:2022-04-01
发布日期:2022-04-18
通讯作者:
沈青春,秦爱建
作者简介:耿仁浩,E-mail: 基金资助:
GENG RenHao1,2( ),LIU Bo1,WANG Fang1,LUO YuFeng1,QU HongFei1,FAN XueZheng1,QIN YuMing1,DING JiaBo1,XU GuanLong1,SHEN QingChun1(
),LIU Bo1,WANG Fang1,LUO YuFeng1,QU HongFei1,FAN XueZheng1,QIN YuMing1,DING JiaBo1,XU GuanLong1,SHEN QingChun1( ),QIN AiJian2(
),QIN AiJian2( )
)
Received:2021-02-08
Accepted:2021-06-21
Online:2022-04-01
Published:2022-04-18
Contact:
QingChun SHEN,AiJian QIN
摘要:
【目的】在生物学研究及生物制品生产中易发生支原体污染,针对我国当前支原体检验方法在时效性和敏感性上的不足,建立一种简便快速、特异敏感的支原体检验PCR方法。【方法】从SILVA数据库下载包含全部细菌、古菌和真菌的核糖体rRNA小亚基(16S/18S, SSU)参考序列的库文件SILVA_123_SSURef,从中提取全部支原体序列,经去重复后得到181条(种)支原体(包括139个已分类的单一种类和42个未分类的支原体)的16S rRNA序列,经比对后选取高变区V6到V9为检测目标区段,设计筛选出表现最佳的检测引物,建立检测支原体的通用PCR方法。选取12种常见的支原体或2种甾原体作为待检样品对该PCR方法进行检测范围验证;取6种不同动物来源的常用传代细胞和3种常见细菌进行特异性验证;选取了5种最常见的支原体和1种甾原体进行敏感性试验;并将其与经典培养法一同对17批(种)动物病毒活疫苗(分别用于6种动物)和24份8种细胞培养物样本进行对比检测以评价其实际应用效果。【结果】建立了一种通用的支原体PCR检测方法,检测引物由2条上游引物(5′-GCAAARCTATRGARAYATAGYVGAG-3′和5′-GCAAAGGCTTAGAAATAAGTTCGGAG-3′)和1条下游引物(5′- CCARCTCYCATRGTKTGACGG-3′)组成,引物配比为3﹕1﹕4,最佳退火温度为56℃。采用该PCR方法对14个较常见的支原体种类进行检测,结果均能扩增出396—413 bp大小的特异性条带,表明该方法的检测范围符合检测要求;对6种动物传代细胞和3种常见细菌进行检测,结果均未扩增出特异性条带,表明其特异性良好;灵敏度测定结果表明该PCR方法的检测下限可达20—200 CCU的活菌,对应的核酸为1.5—15.0 pg;对共计17批病毒活疫苗和24份细胞样品的检测结果与培养法检测结果基本一致,表明本研究建立的支原体PCR检测方法与培养法有很好的一致性,而且敏感性更高。【结论】建立的支原体PCR检测方法特异性强、敏感性高、重复性好、简单快速,为细胞及病毒活疫苗中可能的支原体污染提供了一种准确可靠的快速检测方法。
耿仁浩,刘博,王芳,罗玉峰,曲鸿飞,范学政,秦玉明,丁家波,许冠龙,沈青春,秦爱建. 细胞和病毒活疫苗中支原体污染的PCR检测方法建立与应用[J]. 中国农业科学, 2022, 55(7): 1458-1468.
GENG RenHao,LIU Bo,WANG Fang,LUO YuFeng,QU HongFei,FAN XueZheng,QIN YuMing,DING JiaBo,XU GuanLong,SHEN QingChun,QIN AiJian. Establishment and Application of PCR Assay for Mycoplasma Contamination in Cell Culture and Live Virus Vaccine[J]. Scientia Agricultura Sinica, 2022, 55(7): 1458-1468.
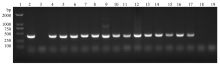
图2
支原体PCR检测引物验证 1:DL2000 Marker;2:阳性对照;3:阴性对照;4:猪鼻支原体(CVCC361);5:鸡滑液支原体(CVCC385);6:鸡毒支原体(CVCC353);7:牛支原体XBY01株;8:猪肺炎支原体(CVCC4049);9:衣阿华支原体(CVCC364);10:人型支原体HB1株;11:鸭支原体BSJ52株;12:绵羊肺炎支原体(CVCC380);13:惰性支原体(CVCC363);14:精氨酸支原体(CVCC346);15:莱氏无胆甾原体(CVCC2406);16:口腔支原体(CVCC379);17:无黄无胆甾原体338株;18:大肠杆菌DH5α;19:Mac145细胞"
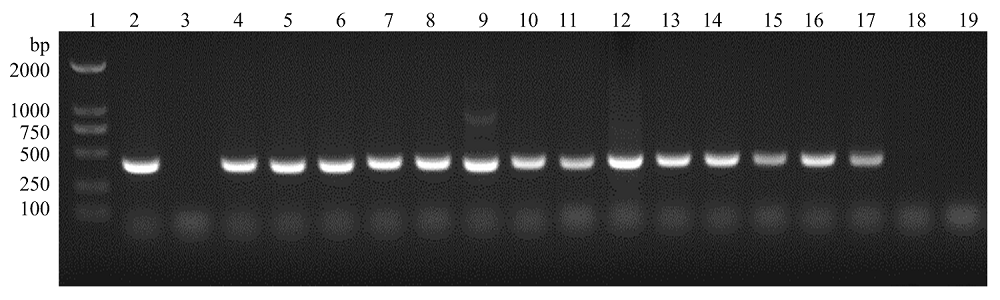
表1
采用支原体检验PCR方法和传统培养法对检测兽用活疫苗的检测"
| 类别 category | 生物制品名称 Biological products | 原材料 Materials | 阳性结果数Number of positive samples | |
|---|---|---|---|---|
| PCR检测 PCR detection | 培养法检测Cultivationa | |||
| 细胞 Cells | 健康细胞8株(种) 8 strains(8 species) of healthy cell | / | 0/8 | 0/8 |
| 待检细胞16株(8种) 16 strains(8 species) of cell samples | / | 6/16 | 4/16(6/16) | |
| 动物用病毒 活疫苗 Live vaccines for animals | 鸡用病毒活疫苗6批(品种) 6 batches (species) of live vaccines for chicken | 鸡胚、CEF等 Chicken embryos, CEF, etc. | 1/6 | 0/6 |
| 鸭用病毒活疫苗2批(品种) 2 batches (species) of live vaccines for duck | 鸭胚 Duck embryos | 0/2 | 0/2 | |
| 鹅用病毒活疫苗1批(品种) 1 batch (species) of live vaccine for geese | 鸭胚 Duck embryos | 0/1 | 0/1 | |
| 猪用病毒活疫苗5批(品种) 3 batches (species) of live vaccines for swine | Vero、PK15、Marc145、ST等 Vero, PK15, Marc145, ST, etc. | 0/5 | 0/5 | |
| 犬用病毒活疫苗2批(品种) 2 batches (species) of live vaccines for dog | 鸡胚、Vero、MDCK、PK15等 Chicken embryos, Vero,MDCK, PK15, etc. | 0/2 | 0/2 | |
| 草鱼用病毒活疫苗1批(品种) 1 batch (species) of live vaccines for grass carp | 草鱼胚胎细胞 Embryonic cell of grass carp | 0/1 | 0/1 | |
| [1] |
UPHOFF C C, DREXLER H G. Detection of mycoplasma contaminations. Methods in Molecular Biology (Clifton, N J), 2013, 946:1-13. doi: 10.1007/978-1-62703-128-8_1.
doi: 10.1007/978-1-62703-128-8_1 |
| [2] |
MEDVEDEVA E S, BARANOVA N B, MOUZYKANTOV A A, GRIGORIEVA T Y, DAVYDOVA M N, TRUSHIN M V, CHERNOVA O A, CHERNOV V M. Adaptation of mycoplasmas to antimicrobial agents: Acholeplasma laidlawii extracellular vesicles mediate the export of ciprofloxacin and a mutant gene related to the antibiotic target. The Scientific World Journal, 2014, 2014:150615. doi: 10.1155/2014/150615.
doi: 10.1155/2014/150615 |
| [3] |
TEYSSOU R, POUTIERS F, SAILLARD C, GRAU O, LAIGRET F, BOVÉ J M, BÉBÉAR C. Detection of mollicute contamination in cell cultures by 16S rDNA amplification. Molecular and Cellular Probes, 1993, 7(3):209-216. doi: 10.1006/mcpr.1993.1030.
doi: 10.1006/mcpr.1993.1030 |
| [4] |
ROBINSON L B, WICHELHAUSEN R H, ROIZMAN B. Contamination of human cell cultures by pleuropneumonialike organisms. Science, 1956, 124(3232):1147-1148. doi: 10.1126/science.124.3232.1147.
doi: 10.1126/science.124.3232.1147 pmid: 13380429 |
| [5] |
VOLOKHOV D V, GRAHAM L J, BRORSON K A, CHIZHIKOV V E. Mycoplasma testing of cell substrates and biologics: Review of alternative non-microbiological techniques. Molecular and Cellular Probes, 2011, 25(2/3):69-77. doi: 10.1016/j.mcp.2011.01.002.
doi: 10.1016/j.mcp.2011.01.002 |
| [6] |
赵翔, 冯建平, 孟淑芳. 支原体检查的核酸检测方法及方法学验证的思考. 中国药事, 2018, 32(8):1020-1027. doi: 10.16153/j.1002-7777.2018.08.003.
doi: 10.16153/j.1002-7777.2018.08.003 |
|
ZHAO X, FENG J P, MENG S F. Considerations on Mycoplasma detection by nucleic acid detection method and the methodological validation. Chinese Pharmaceutical Affairs, 2018, 32(8):1020-1027. doi: 10.16153/j.1002-7777.2018.08.003. (in Chinese)
doi: 10.16153/j.1002-7777.2018.08.003 |
|
| [7] |
ROTTEM S, BARILE M F. Beware of mycoplasmas. Trends in Biotechnology, 1993, 11(4):143-151. doi: 10.1016/0167-7799(93)90089-R.
doi: 10.1016/0167-7799(93)90089-R |
| [8] | 中国兽药典委员会. 中华人民共和国兽药典-二部: 2015年版. 北京: 中国农业出版社, 2016. |
| Chinese Veterinary Pharmacopoeia Commission. Chinese Veterinary Pharmacopoeia of the People's Repubilic of China. Vol 2015. Beijing: Chinese Agriculture Press, 2016. (in Chinese) | |
| [9] | 国家药典委员会. 中华人民共和国药典-一部: 2020年版. 北京: 中国医药科技出版社, 2020. |
| Chinese Pharmacopoeia Commission. Chinese Pharmacopoeia of the People's Repubilic of China. Vol 2020. The Medicine Science and Technology Press of China, 2020. (in Chinese) | |
| [10] |
DREXLER H G, UPHOFF C C. Mycoplasma contamination of cell cultures. Encyclopedia of Cell Technology, 2003. doi: 10.1002/0471250570.spi054.
doi: 10.1002/0471250570.spi054 |
| [11] |
YOUNG L, SUNG J, STACEY G, MASTERS J R. Detection of Mycoplasma in cell cultures. Nature Protocols, 2010, 5(5):929-934. doi: 10.1038/nprot.2010.43.
doi: 10.1038/nprot.2010.43 |
| [12] |
LIGASOVÁ A, VYDRŽALOVÁ M, BURIÁNOVÁ R, BRŮČKOVÁ L, VEČEŘOVÁ R, JANOŠŤÁKOVÁ A, KOBERNA K. A new sensitive method for the detection of mycoplasmas using fluorescence microscopy. Cells, 2019, 8(12):1510. doi: 10.3390/cells8121510.
doi: 10.3390/cells8121510 |
| [13] |
BAI Y, GAN Y, HUA L Z, NATHUES H, YANG H, WEI Y N, WU M, SHAO G Q, FENG Z X. Application of a sIgA-ELISA method for differentiation of Mycoplasma hyopneumoniae infected from vaccinated pigs. Veterinary Microbiology, 2018, 223:86-92. doi: 10.1016/j.vetmic.2018.07.023.
doi: 10.1016/j.vetmic.2018.07.023 |
| [14] |
QASEM J A, AL-MOUQATI S A, AL-ALI E M, BEN-HAJI A. Application of molecular and serological methods for rapid detection of Mycoplasma gallisepticum infection (Avian mycoplasmosis). Pakistan Journal of Biological Sciences: PJBS, 2015, 18(2):81-87. doi: 10.3923/pjbs.2015.81.87.
doi: 10.3923/pjbs.2015.81.87 |
| [15] |
BEN ABDELMOUMEN MARDASSI B, BÉJAOUI A A, OUSSAEIF L, MLIK B, AMOUNA F. A recombinant antigen-based ELISA for the simultaneous differential serodiagnosis of Mycoplasma gallisepticum, Mycoplasma synoviae, and Mycoplasma meleagridis infections. Avian Diseases, 2008, 52(2):214-221. doi: 10.1637/8071-071207-Reg.1.
doi: 10.1637/8071-071207-Reg.1 |
| [16] |
JOHANSSON K E, JOHANSSON I, GÖBEL U B. Evaluation of different hybridization procedures for the detection of mycoplasma contamination in cell cultures. Molecular and Cellular Probes, 1990, 4(1):33-42. doi: 10.1016/0890-8508(90)90037-Z.
doi: 10.1016/0890-8508(90)90037-Z |
| [17] |
KONG H, VOLOKHOV D V, GEORGE J, IKONOMI P, CHANDLER D, ANDERSON C, CHIZHIKOV V. Application of cell culture enrichment for improving the sensitivity of mycoplasma detection methods based on nucleic acid amplification technology (NAT). Applied Microbiology and Biotechnology, 2007, 77(1):223-232. doi: 10.1007/s00253-007-1135-1.
doi: 10.1007/s00253-007-1135-1 |
| [18] |
UPHOFF C C, DREXLER H G. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods in Molecular Biology (Clifton, N J), 2011, 731:93-103. doi: 10.1007/978-1-61779-080-5_8.
doi: 10.1007/978-1-61779-080-5_8 |
| [19] |
英聪, 王海光, 刘灿, 沈青春, 毕丁仁, 宁宜宝. 检测污染兽用疫苗14种支原体PCR方法的建立及应用. 中国预防兽医学报, 2014, 36(1):42-45. doi: 10.3969/j.issn.1008-0589.2014.01.11.
doi: 10.3969/j.issn.1008-0589.2014.01.11 |
|
YING C, WANG H G, LIU C, SHEN Q C, BI D R, NING Y B. The establishment of the PCR assay for detection of 14 kinds of Mycoplasma contamination in veterinary vaccines. Chinese Journal of Preventive Veterinary Medicine, 2014, 36(1):42-45. doi: 10.3969/j.issn.1008-0589.2014.01.11. (in Chinese)
doi: 10.3969/j.issn.1008-0589.2014.01.11 |
|
| [20] |
INGEBRITSON A L, GIBBS C P, TONG C, SRINIVAS G B. A PCR detection method for testing mycoplasma contamination of veterinary vaccines and biological products. Letters in Applied Microbiology, 2015, 60(2):174-180. doi: 10.1111/lam.12355.
doi: 10.1111/lam.12355 |
| [21] |
MBELO S, GAY V, BLANCHARD S, ABACHIN E, FALQUE S, LECHENET J, POULET H, DE SAINT-VIS B. Development of a highly sensitive PCR/DNA chip method to detect mycoplasmas in a veterinary modified live vaccine. Biologicals, 2018, 54:22-27. doi: 10.1016/j.biologicals.2018.05.002.
doi: 10.1016/j.biologicals.2018.05.002 |
| [22] |
高广仁, 万玉林, 崔治亮, 葛玉凤, 刘学荣. 兽用生物制品中支原体污染PCR检测方法的建立与应用. 黑龙江畜牧兽医, 2018(15):163-166. doi: 10.13881/j.cnki.hljxmsy.2017.12.0345.
doi: 10.13881/j.cnki.hljxmsy.2017.12.0345 |
|
GAO G R, WAN Y L, CUI Z L, GE Y F, LIU X R. Establishment and application of PCR for detection of mycoplasma contamination in veterinary biological products. Heilongjiang Animal Science and Veterinary Medicine, 2018(15):163-166. doi: 10.13881/j.cnki.hljxmsy.2017.12.0345. (in Chinese)
doi: 10.13881/j.cnki.hljxmsy.2017.12.0345 |
|
| [23] |
ELDERING J A, FELTEN C, VEILLEUX C A, POTTS B J. Development of a PCR method for mycoplasma testing of Chinese hamster ovary cell cultures used in the manufacture of recombinant therapeutic proteins. Biologicals, 2004, 32(4):183-193. doi: 10.1016/j.biologicals.2004.08.005.
doi: 10.1016/j.biologicals.2004.08.005 |
| [24] |
GRÓZNER D, SULYOK K M, KREIZINGER Z, RÓNAI Z, JÁNOSI S, TURCSÁNYI I, KÁROLYI H F, KOVÁCS Á B, KISS M J, VOLOKHOV D, GYURANECZ M. Detection of Mycoplasma anatis, M. anseris, M. cloacale and Mycoplasma sp. 1220 in waterfowl using species-specific PCR assays. PLoS ONE, 2019, 14(7):e0219071. doi: 10.1371/journal.pone.0219071.
doi: 10.1371/journal.pone.0219071 |
| [25] |
WISSELINK H J, SMID B, PLATER J, RIDLEY A, ANDERSSON A M, ASPÁN A, POHJANVIRTA T, VÄHÄNIKKILÄ N, LARSEN H, HØGBERG J, COLIN A, TARDY F. A European interlaboratory trial to evaluate the performance of different PCR methods for Mycoplasma bovis diagnosis. BMC Veterinary Research, 2019, 15(1):86. doi: 10.1186/s12917-019-1819-7.
doi: 10.1186/s12917-019-1819-7 |
| [26] |
TOTTEN A H, LEAL S M Jr, RATLIFF A E, XIAO L, CRABB D M, WAITES K B. Evaluation of the ELITe InGenius PCR platform for detection of Mycoplasma pneumoniae. Journal of Clinical Microbiology, 2019, 57(6):e00287-e00219. doi: 10.1128/JCM.00287-19.
doi: 10.1128/JCM.00287-19 |
| [27] |
JEAN A, TARDY F, ALLATIF O, GROSJEAN I, BLANQUIER B, GERLIER D. Assessing mycoplasma contamination of cell cultures by qPCR using a set of universal primer pairs targeting a 1.5 kb fragment of 16S rRNA genes. PLoS ONE, 2017, 12(2):e0172358. doi: 10.1371/journal.pone.0172358.
doi: 10.1371/journal.pone.0172358 |
| [28] |
BASCUÑANA C R, MATTSSON J G, BÖLSKE G, JOHANSSON K E. Characterization of the 16S rRNA genes from Mycoplasma sp. strain F38 and development of an identification system based on PCR. Journal of Bacteriology, 1994, 176(9):2577-2586. doi: 10.1128/jb.176.9.2577-2586.1994.
doi: 10.1128/jb.176.9.2577-2586.1994 |
| [29] |
AMRAM E, MIKULA I, SCHNEE C, AYLING R D, NICHOLAS R A J, ROSALES R S, HARRUS S, LYSNYANSKY I. 16S rRNA gene mutations associated with decreased susceptibility to tetracycline in Mycoplasma bovis. Antimicrobial Agents and Chemotherapy, 2015, 59(2):796-802. doi: 10.1128/AAC.03876-14.
doi: 10.1128/AAC.03876-14 |
| [30] |
BAKER G C, SMITH J J, COWAN D A. Review and re-analysis of domain-specific 16S primers. Journal of Microbiological Methods, 2003, 55(3):541-555. doi: 10.1016/j.mimet.2003.08.009.
doi: 10.1016/j.mimet.2003.08.009 |
| [31] |
BAHRAM M, ANSLAN S, HILDEBRAND F, BORK P, TEDERSOO L. Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environmental Microbiology Reports, 2019, 11(4):487-494. doi: 10.1111/1758-2229.12684.
doi: 10.1111/1758-2229.12684 |
| [32] |
代玉立, 甘林, 滕振勇, 杨静民, 祁月月, 石妞妞, 陈福如, 杨秀娟. 玉米大斑病菌和小斑病菌交配型多重PCR检测方法的建立与应用. 中国农业科学, 2020, 53(3):527-538. doi: 10.3864/j.issn.0578-1752.2020.03.006.
doi: 10.3864/j.issn.0578-1752.2020.03.006 |
|
DAI Y L, GAN L, TENG Z Y, YANG J M, QI Y Y, SHI N N, CHEN F R, YANG X J. Establishment and application of a multiple PCR method to detect mating types of Exserohilum turcicum and Bipolaris maydis. Scientia Agricultura Sinica, 2020, 53(3):527-538. doi: 10.3864/j.issn.0578-1752.2020.03.006. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2020.03.006 |
|
| [33] |
文静, 郭勇, 邱丽娟. 耐草甘膦转EPSPS/GAT大豆多重PCR检测体系的建立及应用. 中国农业科学, 2020, 53(20):4127-4136. doi: 10.3864/j.issn.0578-1752.2020.20.003.
doi: 10.3864/j.issn.0578-1752.2020.20.003 |
|
WEN J, GUO Y, QIU L J. Establishment and application of multiple PCR detection system for glyphosate-tolerant gene EPSPS/GAT in soybean. Scientia Agricultura Sinica, 2020, 53(20):4127-4136. doi: 10.3864/j.issn.0578-1752.2020.20.003. (in Chinese)
doi: 10.3864/j.issn.0578-1752.2020.20.003 |
|
| [34] | NIKFARJAM L, FARZANEH P. Prevention and detection of Mycoplasma contamination in cell culture. Cell Journal, 2012, 13(4):203-212. |
| [35] |
UPHOFF C C, DREXLER H G. Eradication of Mycoplasma contaminations from cell cultures. Current Protocols in Molecular Biology, 2014, 106:28.5.1-28.512. doi: 10.1002/0471142727.mb2805s106.
doi: 10.1002/0471142727.mb2805s106 |
| [36] |
UPHOFF C C, DREXLER H G. Eradication of Mycoplasma contaminations. Basic Cell Culture Protocols, 2013, 946:15-26. doi: 10.1007/978-1-62703-128-8_2.
doi: 10.1007/978-1-62703-128-8_2 |
| [37] | UPHOFF C C, DREXLER H G. Detection of mycoplasma contaminations in cell cultures by PCR analysis. Human Cell, 1999, 12(4):229-236. |
| [38] |
KONG F, JAMES G, GORDON S, ZELYNSKI A, GILBERT G L. Species-specific PCR for identification of common contaminant mollicutes in cell culture. Applied and Environmental Microbiology, 2001, 67(7):3195-3200. doi: 10.1128/AEM.67.7.3195-3200.2001.
doi: 10.1128/AEM.67.7.3195-3200.2001 |
| [39] |
钱利武, 罗京京, 王浩, 刘小琼, 王慧. 药品GMP检查中质量控制与质量保证方面存在的主要问题及建议. 中国药事, 2020, 34(1):17-21. doi: 10.16153/j.1002-7777.2020.01.003.
doi: 10.16153/j.1002-7777.2020.01.003 |
|
QIAN L W, LUO J J, WANG H, LIU X Q, WANG H. Main problems of quality control and quality assurance in drug GMP inspection and the suggestions. Chinese Pharmaceutical Affairs, 2020, 34(1):17-21. doi: 10.16153/j.1002-7777.2020.01.003. (in Chinese)
doi: 10.16153/j.1002-7777.2020.01.003 |
|
| [40] |
谢寅, 刘晓萌, 吕建军, 孟建华, 王秀文. 瑞士GLP法令与中国GLP规范的主要差异. 中国药事, 2016, 30(4):352-354. doi: 10.16153/j.1002-7777.2016.04.008.
doi: 10.16153/j.1002-7777.2016.04.008 |
|
XIE Y, LIU X M, LÜ J J, MENG J H, WANG X W. Major differences between Swiss GLP articles and Chinese GLP. Chinese Pharmaceutical Affairs, 2016, 30(4):352-354. doi: 10.16153/j.1002-7777.2016.04.008. (in Chinese)
doi: 10.16153/j.1002-7777.2016.04.008 |
| [1] | 李菲菲, 廉雪菲, 尹韬, 常媛媛, 金燕, 马小川, 陈岳文, 叶丽, 李云松, 卢晓鹏. 柑橘果实囊衣发育与化渣性的形成[J]. 中国农业科学, 2023, 56(2): 333-344. |
| [2] | 李旭飞,杨盛迪,李松琦,刘海楠,裴茂松,韦同路,郭大龙,余义和. 葡萄VlCKX4表达特性分析与转录调控预测[J]. 中国农业科学, 2023, 56(1): 144-155. |
| [3] | 翟晓虎,李翎旭,陈小竹,蒋怀德,贺卫华,姚大伟. 肉中猪源性成分Real-time PCR定量检测技术[J]. 中国农业科学, 2023, 56(1): 156-164. |
| [4] | 杨昕冉,马鑫浩,杜嘉伟,昝林森. m6A甲基化酶相关基因在牛骨骼肌生成中的表达[J]. 中国农业科学, 2023, 56(1): 165-178. |
| [5] | 王一丹,杨发龙,陈弟诗,向华,任玉鹏. 猪腹泻病毒一步法多重TaqMan荧光定量RT-PCR检测法的建立及应用[J]. 中国农业科学, 2023, 56(1): 179-192. |
| [6] | 刘玉芳,陈玉林,周祖阳,储明星. miR-221-3p靶向BCL2L11调控小尾寒羊卵泡颗粒细胞凋亡[J]. 中国农业科学, 2022, 55(9): 1868-1876. |
| [7] | 王思彤,陈艳,罗雨嘉,杨缘缘,蒋志洋,蒋鑫怡,钟樊,陈好,徐红星,吴俨,段红霞,唐斌. 三种新型化合物对草地贪夜蛾海藻糖与几丁质代谢及生长发育的影响[J]. 中国农业科学, 2022, 55(8): 1568-1578. |
| [8] | 吴艳,张昊,梁振华,潘爱銮,申杰,蒲跃进,黄涛,皮劲松,杜金平. circ-13267通过let-7-19/ERBB4通路调控蛋鸭卵泡颗粒细胞凋亡[J]. 中国农业科学, 2022, 55(8): 1657-1666. |
| [9] | 阿依木古丽·阿不都热依木,阿尔祖古丽·阿依丁,王家敏,石嘉琛,马芳芳,蔡勇,乔自林. 大豆异黄酮对牦牛卵巢颗粒细胞增殖和凋亡的影响[J]. 中国农业科学, 2022, 55(8): 1667-1675. |
| [10] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [11] | 彭雪,高月霞,张琳煊,高志强,任亚梅. 高能电子束辐照对马铃薯贮藏品质及芽眼细胞超微结构的影响[J]. 中国农业科学, 2022, 55(7): 1423-1432. |
| [12] | 李文慧,贺依静,姜瑶,赵红宇,彭磊,李佳,芮荣,剧世强. 伏马毒素B1对猪体外成熟卵母细胞凋亡与自噬的影响[J]. 中国农业科学, 2022, 55(6): 1241-1252. |
| [13] | 赫磊,路凯,赵春芳,姚姝,周丽慧,赵凌,陈涛,朱镇,赵庆勇,梁文化,王才林,朱丽,张亚东. 水稻穗顶端退化突变体paa21的表型分析及基因克隆[J]. 中国农业科学, 2022, 55(24): 4781-4792. |
| [14] | 郭绍雷,许建兰,王晓俊,宿子文,张斌斌,马瑞娟,俞明亮. 桃XTH家族基因鉴定及其在桃果实贮藏过程中的表达特性[J]. 中国农业科学, 2022, 55(23): 4702-4716. |
| [15] | 谢丽雪,张小艳,张立杰,郑姗,李韬. 侵染西番莲的东亚西番莲病毒全基因组序列特征及TC-RT-PCR检测技术[J]. 中国农业科学, 2022, 55(22): 4408-4418. |
|
||