













中国农业科学 ›› 2024, Vol. 57 ›› Issue (3): 442-453.doi: 10.3864/j.issn.0578-1752.2024.03.002
熊楚雯( ), 郭智滨(
), 郭智滨( ), 周强华, 程艳波, 马启彬, 蔡占东, 年海(
), 周强华, 程艳波, 马启彬, 蔡占东, 年海( )
)
收稿日期:2023-06-14
接受日期:2023-07-28
出版日期:2024-02-01
发布日期:2024-02-05
通信作者:
联系方式:
熊楚雯,E-mail:xiongchuwen3222@163.com。郭智滨,E-mail:20222015014@stu.scau.edu.cn。熊楚雯和郭智滨为同等贡献者。
基金资助:
XIONG ChuWen( ), GUO ZhiBin(
), GUO ZhiBin( ), ZHOU QiangHua, CHENG YanBo, MA QiBin, CAI ZhanDong, NIAN Hai(
), ZHOU QiangHua, CHENG YanBo, MA QiBin, CAI ZhanDong, NIAN Hai( )
)
Received:2023-06-14
Accepted:2023-07-28
Published:2024-02-01
Online:2024-02-05
摘要:
【目的】磷含量偏低是影响酸性土壤作物产量的重要因素。大豆(Glycine max)是重要的粮食和油料作物,也为喜磷作物,缺磷则影响其产量与品质。NAC(NAM,ATAF1/2,CUC2)转录因子家族参与多种植物对生物胁迫和非生物胁迫响应的调控,是否参与大豆低磷胁迫响应尚未深入研究。以耐低磷野生大豆BW69为材料,克隆获得耐低磷基因GsNAC1并对其表达特性及功能进行分析,为深入解析GsNAC1调控大豆低磷胁迫及其机制奠定基础。【方法】从野生大豆BW69克隆GsNAC1的全长序列,并通过生物信息学分析探究其编码氨基酸序列的特征。随后,利用实时荧光定量PCR技术(qRT-PCR)对其组织表达模式进行分析,并通过激光共聚焦显微镜观察其编码蛋白的亚细胞定位。此外,通过大豆遗传转化试验,获得转基因株系并进行表型分析。最后,通过转录组联合分析来鉴定转基因植株中与低磷胁迫相关的差异表达基因(differentially expressed genes,DEGs)。【结果】成功克隆获得GsNAC1,编码区全长876 bp,通过构建系统发育树发现GsNAC1与AtATAF1的序列相似性为62.46%,与Williams 82参考基因组的GmNAC1序列没有差异;进一步的亚细胞定位结果显示,GsNAC1定位于细胞核;基于qRT-PCR技术,发现GsNAC1在大豆的根、茎、叶、顶端、花和豆荚均有表达,在根部的相对表达量最高,且受到低pH和低磷诱导表达显著上调。通过水培法和土培法进行表型试验,在低磷处理下,与野生型(WT)相比,转基因株系鲜重根冠比、总根长、根表面积、根体积和磷含量均显著高于WT。结合转录组测序数据进行分析,发现GsNAC1可能通过促进GmALMT6、GmALMT27、GmPAP27和GmWRKY21等基因表达增强其对低磷胁迫的耐受性。【结论】GsNAC1受低pH和低磷诱导表达上调,过量表达GsNAC1可以显著增强大豆对低磷胁迫的耐受性,在低磷胁迫反应中起促进作用。GsNAC1可能通过调控下游基因表达增强大豆对酸性低磷胁迫的耐受性。
熊楚雯, 郭智滨, 周强华, 程艳波, 马启彬, 蔡占东, 年海. 大豆转录因子NAC1耐低磷胁迫的功能研究[J]. 中国农业科学, 2024, 57(3): 442-453.
XIONG ChuWen, GUO ZhiBin, ZHOU QiangHua, CHENG YanBo, MA QiBin, CAI ZhanDong, NIAN Hai. Function Analysis of the Soybean Transcription Factor NAC1 in Tolerance to Low Phosphorus[J]. Scientia Agricultura Sinica, 2024, 57(3): 442-453.
表1
试验所用引物序列"
| 引物名称 Primer name | 序列 Sequences (5′-3′) |
|---|---|
| GsNAC1-F | ACGTTTCAAATCGCAGTCGC |
| GsNAC1-R | CTTGGCCTAGCCCACATCAC |
| PTF101-GsNAC1-F | gagaacacgggggactctagaATGAAGGGAGAATTAGAGTTGCCA |
| PTF101-GsNAC1-R | cgatcggggaaattcgagctcTCACATCTTCTGTAGGTACATGAACATG |
| GsNAC1-GFP-F | acgggggactcttgaccatggATGAAGGGAGAATTAGAGTTGCCA |
| GsNAC1-GFP-R | aagttcttctcctttactagtCCATCTTCTGTAGGTACATGAACATG |
| RT-GsNAC1-F | CGATCGGAAAACCGAAAGCG |
| RT-GsNAC1-R | TCAACATTGGCGAGGCGATA |
| Actin3-F | GCACCACCGGAGAGAAAATA |
| Actin3-R | GTGCACAATTGATGGACCAG |
| qGmALMT6-F | CCTTGAAGCATCAAAAGAGCCCA |
| qGmALMT6-R | CCCATTGTGACTGCCTCAAAGAT |
| qGmALMT27-F | TTTGAGTTTACCGCAGGGGC |
| qGmALMT27-R | TACGCGGTCAGATCCATTGTC |
| qGmPAP27-F | ACATGTCCTGACACTGACGG |
| qGmPAP27-R | TTGCCACAAGTCTAGGATCGG |
| qGmWRKY21-F | ACAATCCCAACCCAAGGAACT |
| qGmWRKY21-R | GCACCAAGGGAATTTGGCTG |
| 35S | CGGATTCCATTGCCCAGCTA |
| NOS-R | CACCGCGCGCGATAATTT |
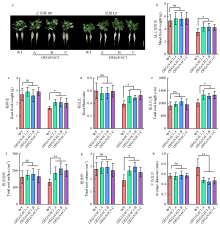
图3
在LP和NP处理下WT和转基因株系的长期水培表型鉴定 a:WT和转基因株系在长期水培试验中的生长情况。利用RhizoVision Explorer软件分析处理15 d后的根系地上部鲜重(b)、根鲜重(c)、根冠比(d)、根总长度(e)、根表面积(f)、根体积(g)和平均直径(h)的形态参数。WT:野生型(华春6号),转基因株系:OEGsNAC1-A、OEGsNAC1-B和OEGsNAC1-C,A:OEGsNAC1-A,B:OEGsNAC1-B,C:OEGsNAC1-C。ns:表示不显著,*表示在P<0.05水平差异显著,**表示在P<0.01水平差异极显著。数据以3个生物重复的均值±标准差表示。下同"

| [1] |
doi: S1369-5266(14)00096-X pmid: 25036899 |
| [2] |
沈启维, 李艳春, 张健. 提高植物磷高效利用能力方法的研究进展. 绿色科技, 2021, 23(11): 157-160.
|
|
|
|
| [3] |
pmid: 15377225 |
| [4] |
doi: 10.1016/j.cj.2019.11.002 |
| [5] |
刘建中, 李振声, 李继云. 利用植物自身潜力提高土壤中磷的生物有效性. 生态农业研究, 1994, 2(1): 16-23.
|
|
|
|
| [6] |
|
| [7] |
doi: 10.1111/nph.2013.198.issue-3 |
| [8] |
doi: 10.1371/journal.pgen.1006194 |
| [9] |
doi: 10.1093/plphys/kiad170 |
| [10] |
doi: 10.1104/pp.113.235077 |
| [11] |
doi: 10.3390/ijms232315274 |
| [12] |
doi: 10.3389/fpls.2015.00902 pmid: 26579152 |
| [13] |
|
| [14] |
doi: 10.1371/journal.pone.0084886 |
| [15] |
doi: 10.1111/tpj.2011.68.issue-2 |
| [16] |
doi: 10.3390/ijms232012378 |
| [17] |
|
| [18] |
李雅雪, 盘耀亮, 彭光粉, 田江, 陆星, 梁翠月. GmNTLs调控大豆根系响应低磷胁迫的功能研究. 华南农业大学学报, 2023, 44(2): 221-229.
|
|
|
|
| [19] |
doi: 10.1006/meth.2001.1262 |
| [20] |
鲍士旦. 土壤农化分析. 3版. 北京: 中国农业出版社, 2000: 268-270.
|
|
|
|
| [21] |
doi: 10.1093/bioinformatics/btp336 pmid: 19497933 |
| [22] |
doi: 10.1038/nmeth.1226 pmid: 18516045 |
| [23] |
doi: 10.1111/jipb.v60.3 |
| [24] |
doi: 10.3389/fpls.2020.00661 |
| [25] |
doi: 10.1007/s12892-019-0102-0 |
| [26] |
doi: 10.1038/nature11346 |
| [27] |
doi: 10.3390/cells11030525 |
| [28] |
doi: 10.1016/j.tplants.2012.02.004 pmid: 22445067 |
| [29] |
doi: 10.1093/dnares/10.6.239 |
| [30] |
doi: 10.1038/cr.2009.108 |
| [31] |
doi: 10.3389/fpls.2018.01864 |
| [32] |
万会娜, 于月华, 王怡, 倪志勇. 大豆GmNAC131基因的生物信息学及表达分析. 大豆科学, 2021, 40(2): 186-194.
|
|
|
|
| [33] |
严小龙, 廖红, 戈振扬, 罗锡文. 植物根构型特性与磷吸收效率. 植物学通报, 2000, 35(6): 511-519.
|
|
|
|
| [34] |
|
| [35] |
doi: 10.1104/pp.111.173724 pmid: 21464471 |
| [36] |
doi: 10.1146/arplant.2022.73.issue-1 |
| [37] |
|
| [38] |
|
| [39] |
doi: 10.1016/j.tplants.2008.10.004 pmid: 19056309 |
| [40] |
doi: 10.1007/s00299-014-1588-5 |
| [41] |
doi: 10.1016/j.plantsci.2022.111283 |
| [42] |
doi: 10.3389/fpls.2019.01714 |
| [1] | 卫乃翠, 陶金博, 苑名杨, 张彧, 开梦想, 乔玲, 武棒棒, 郝宇琼, 郑兴卫, 王娟玲, 赵佳佳, 郑军. 山西小麦苗期耐低磷特性及遗传分析[J]. 中国农业科学, 2024, 57(5): 831-845. |
| [2] | 姬改革, 陈智武, 单艳菊, 刘一帆, 屠云洁, 邹剑敏, 章明, 巨晓军, 束婧婷, 张海涛, 唐燕飞, 蒋华莲. 基于转录组测序研究调控黄羽肉鸡干毛性状关键基因和信号通路[J]. 中国农业科学, 2024, 57(1): 204-215. |
| [3] | 杨莎, 刘珂珂, 刘颖, 郭峰, 王建国, 高华鑫, 孟静静, 张佳蕾, 万书波. 基于转录组分析花生单粒精播与多粒穴播植株荚果产量差异的分子机制[J]. 中国农业科学, 2023, 56(22): 4386-4402. |
| [4] | 艾代龙, 雷芳, 邹乔生, 贺鹏, 杨洪坤, 樊高琼. 秸秆覆盖与施氮对丘陵旱地小麦根系构型改善及H+、NO3-吸收与利用的影响[J]. 中国农业科学, 2023, 56(21): 4192-4207. |
| [5] | 米热扎提江·喀由木, 西尔艾力·吾麦尔江, 李晓曈, 王香茹, 贵会平, 张恒恒, 张西岭, 董强, 宋美珍. 棉花苗期耐低磷种质筛选及耐低磷综合评价[J]. 中国农业科学, 2023, 56(21): 4150-4162. |
| [6] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [7] | 姜芬芬, 孙磊, 刘方东, 王吴彬, 邢光南, 张焦平, 张逢凯, 李宁, 李艳, 贺建波, 盖钧镒. 世界大豆生育阶段光温综合反应的地理分化和演化[J]. 中国农业科学, 2022, 55(3): 451-466. |
| [8] | 张晓萍,撒世娟,伍涵宇,乔丽媛,郑蕊,姚新灵. 马铃薯叶片气孔的开张与关闭同步伴随果胶的降解与合成[J]. 中国农业科学, 2022, 55(17): 3278-3288. |
| [9] | 解斌,安秀红,陈艳辉,程存刚,康国栋,周江涛,赵德英,李壮,张艳珍,杨安. 不同苹果砧木对持续低磷的响应及适应性评价[J]. 中国农业科学, 2022, 55(13): 2598-2612. |
| [10] | 朱芳芳,董亚辉,任真真,王志勇,苏慧慧,库丽霞,陈彦惠. 过表达ZmIBH1-1提高玉米苗期抗旱性[J]. 中国农业科学, 2021, 54(21): 4500-4513. |
| [11] | 刘锴,何闪闪,张彩霞,张利义,卞书迅,袁高鹏,李武兴,康立群,丛佩华,韩晓蕾. 苹果叶片不定芽再生过程的差异表达基因鉴定与分析[J]. 中国农业科学, 2021, 54(16): 3488-3501. |
| [12] | 张稳,孟淑君,王琪月,万炯,马拴红,林源,丁冬,汤继华. 玉米pTAC2影响苗期叶片叶绿素合成的转录组分析[J]. 中国农业科学, 2020, 53(5): 874-889. |
| [13] | 徐志军, 赵胜, 徐磊, 胡小文, 安东升, 刘洋. 基于RNA-seq数据的栽培种花生SSR位点鉴定和标记开发[J]. 中国农业科学, 2020, 53(4): 695-706. |
| [14] | 高艳,朱雅楠,李秋方,苏松坤,聂红毅. 转录组学分析意大利蜜蜂脑部哺育行为相关基因[J]. 中国农业科学, 2020, 53(19): 4092-4102. |
| [15] | 郭书磊,鲁晓民,齐建双,魏良明,张新,韩小花,岳润清,王振华,铁双贵,陈彦惠. 利用RNA-Seq发掘玉米叶片形态建成相关的调控基因[J]. 中国农业科学, 2020, 53(1): 1-17. |
|
||