













中国农业科学 ›› 2021, Vol. 54 ›› Issue (11): 2273-2286.doi: 10.3864/j.issn.0578-1752.2021.11.003
张林林( ),智慧,汤沙,张仁梁,张伟,贾冠清(
),智慧,汤沙,张仁梁,张伟,贾冠清( ),刁现民(
),刁现民( )
)
收稿日期:2020-11-09
接受日期:2020-12-07
出版日期:2021-06-01
发布日期:2021-06-09
通讯作者:
贾冠清,刁现民
作者简介:张林林,E-mail:基金资助:
ZHANG LinLin( ),ZHI Hui,TANG Sha,ZHANG RenLiang,ZHANG Wei,JIA GuanQing(
),ZHI Hui,TANG Sha,ZHANG RenLiang,ZHANG Wei,JIA GuanQing( ),DIAO XianMin(
),DIAO XianMin( )
)
Received:2020-11-09
Accepted:2020-12-07
Online:2021-06-01
Published:2021-06-09
Contact:
GuanQing JIA,XianMin DIAO
摘要:
【目的】谷子抽穗时间的适应性表现是广适性新品种选育的基础,分析抽穗时间关键基因的遗传变异和单倍型效应,为品种适应性改良提供基础信息。【方法】通过全基因组关联分析(genome-wide association study,GWAS),定位谷子抽穗时间关键基因SiTOC1,利用多组学数据库(multi-omics database for Setaria italica,MDSi)提供的SiTOC1数字表达量,分析SiTOC1的组织时空表达特性,并利用原生质体对SiTOC1蛋白进行亚细胞定位。采用qRT-PCR在短日(10 h光照/14 h黑暗)条件下进行SiTOC1 24 h节律表达模式分析。利用有代表性的99份谷子品种,分析SiTOC1编码区和启动子区的遗传多态性、单倍型以及转录水平,并对单倍型与抽穗时间的关系进行鉴定。【结果】在第1染色体物理位置31 456 761 bp处鉴定到了一个显著的关联信号,与抽穗时间紧密相关,该位点附近存在一个拟南芥抽穗期TOC1的同源基因SiTOC1。SiTOC1在光周期响应组织(根、茎、叶等)中高表达,亚细胞定位于细胞核,在傍晚表达量上调,呈现出24 h节律性表达模式。SiTOC1在不同谷子品种中存在丰富的多态性,但REC和CCT结构域高度保守。SiTOC1编码区2种主要单倍型H-2和H-6分别与启动子单倍型Hp-591C和Hp-591A共分离,其中,启动子单倍型Hp-591C较Hp-591A的相对表达量显著上调了约2.5倍(P=0.014),并且该单倍型在三亚市、长治市和乌鲁木齐市3个环境下的抽穗时间分别平均延迟9、11和12 d。【结论】SiTOC1启动子区第591 bp处的SNP是引起抽穗时间差异的主效位点,单倍型Hp-591A较Hp-591C早熟,可作为主效单倍型用于分子育种选择。
张林林,智慧,汤沙,张仁梁,张伟,贾冠清,刁现民. 谷子抽穗时间基因SiTOC1的表达与单倍型变异分析[J]. 中国农业科学, 2021, 54(11): 2273-2286.
ZHANG LinLin,ZHI Hui,TANG Sha,ZHANG RenLiang,ZHANG Wei,JIA GuanQing,DIAO XianMin. Characterizations of Transcriptional and Haplotypic Variations of SiTOC1 in Foxtail Millet[J]. Scientia Agricultura Sinica, 2021, 54(11): 2273-2286.
表1
SiTOC1亚细胞定位载体构建PCR扩增及测序引物"
| 载体 Vector | 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 产物长度 Product length (bp) |
|---|---|---|---|
| GFP-N 端载体 GFP-N terminal vector | GFPG5-F1 | GCTGTACAAGACTAGTATGGTGGGCGGCGGCG | 1517 |
| GFPG5-R1 | GGGGAAATTCGAGCTCCTACTCTGGAGAAGAAATAATC | ||
| PUCGFPN-TestF | TACAACTACAACAGCCACAA | 2163 | |
| PUCGFPN-TestR | CCTCTTCGCTATTACGC | ||
| GFPG5-F1.1 | CCATCATCATGATGTCC | ||
| GFPG5-R1.1 | TTCAGAGCGACTGCATG | ||
| GFP-C端载体 GFP-C terminal vector | G5GFP-F1 | TAGTGGATCCATCGATATGGTGGGCGGCGGCG | 1514 |
| G5GFP-R1 | TCCCGGGAGCGGTACCCTCTGGAGAAGAAATAATCTC | ||
| PUCGFPC-TestF | ACCTCCTCGGATTCCAT | 2354 | |
| PUCGFPC-TestR | TGCCGTTCTTCTGCTTG | ||
| G5GFP-F1.1 | CAAGATGCTCAAGTACA | ||
| G5GFP-R1.1 | CGATTTACATACCTCACT | ||
| RFP-N端载体 RFP-N terminal vector | MYBRFP-F | GCTGTACAAGACTAGTATGGACATGGCGCACGAGAG | 1022 |
| MYBRFP-R | GGGGAAATTCGAGCTCTCACACGGCGGCCTGGGT | ||
| PUCRFPN-TestF | CTCAAGCTCAAGGACGG | 1584 | |
| PUCRFPN-TestR | CCTCTTCGCTATTACGC |
表2
SiTOC1 PCR扩增和测序引物"
| 产物Product | 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 产物长度 Product length (bp) |
|---|---|---|---|
| 编码区 Coding region | G5-F1 | GATCCAGCGACAGTCCA | 1450 |
| G5-R1 | AGACAGGTCGGACTGAAATA | ||
| G5-F2 | GATGAGTTGCCACAAAGATG | 1272 | |
| G5-R2 | TGTATTGCCTTTCCCAGTAG | ||
| 启动子区 Promoter region | G5P-F1 | GGCCAAACGAAACCATG | 2069 |
| G5P-R1 | CCTAATCCGGGAAACCAG | ||
| G5P-F1.1 | ATTGCGTGCACGAATCT | ||
| G5P-R1.1 | CAGCAGCAGCCTGCCTCG | ||
| SiTOC1互补DNA SiTOC1 cDNA | G5Q-F1 | GCCGATCAAGCATCATATGTTAAGT | 250 |
| G5Q-R1 | TTTGGCCTTCATTGCTTCGC | ||
| Cullin互补DNA Cullin cDNA | Cullin-F | TATGGGTCATCAACAGCTTGTC | 112 |
| Cullin-R | GTAGTCCCTCGTGATGAGATCC |
表3
候选基因注释信息"
| 位点名称 LocusName | 拟南芥对应基因 Arabi-symbol | 拟南芥基因功能注释 Arabi-defline |
|---|---|---|
| Seita.1G235800 | N/A | 无功能注释N/A |
| Seita.1G235900 | MKM21.12 | 线粒体核糖体蛋白L27 Mitochondrial ribosomal protein L27 |
| Seita.1G236000 | N/A | 谷氧还蛋白家族 Glutaredoxin family protein |
| Seita.1G236100 | TOC1 | 含CCT基序的应答调节蛋白 CCT motif-containing response regulator protein |
| Seita.1G236200 | PAP10 | 紫色酸性磷酸酶10 Purple acid phosphatase 10 |
| Seita.1G236300 | NOP10 | 核仁RNA结合Nop10p家族蛋白 Nucleolar RNA-binding Nop10p family protein |
| Seita.1G236400 | AtMYB79 | 含MYB结构域蛋白79 MYB domain protein 79 |
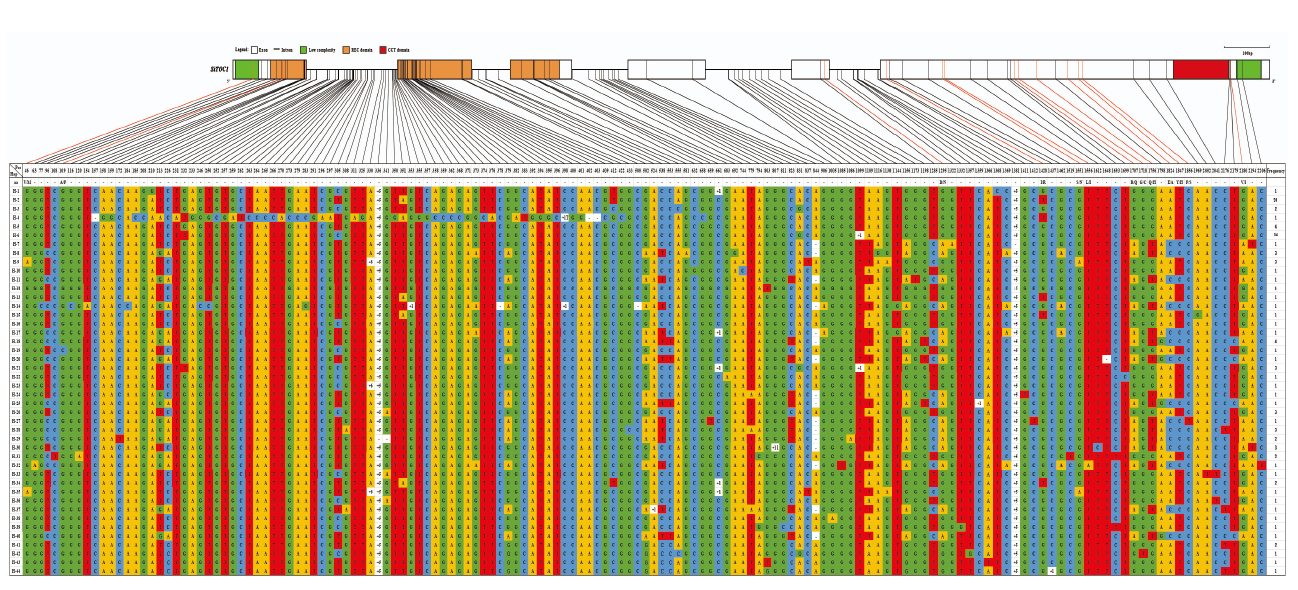
图5
SiTOC1编码区结构变异 图上方为SiTOC1编码区基因结构,其中,框格代表外显子,框格之间的连接线代表内含子,下方对应不同单倍型组合信息表,连接上方SiTOC1编码区基因结构图的实心线指示变异位点,其中,红色连接线指示非同义突变,表格中不同颜色单元格代表不同碱基,其中,黄色、红色、蓝色和绿色单元格分别代表A碱基、T碱基、C碱基、G碱基,-:缺失,+:插入,符号后面的数字代表缺失或插入碱基数,Indel位点从左往右依次为:330(+6)/+gtgctg、336(+5)/+atgct、353(+1)/+t、398(+1)/+t、524(+1)/+t、661(+1)/+g、807(+11)/+gtgggttgctt、1099(+1)/+c、1359(+1)/+t、1381(+5)/+taatc、1437(+1)/+t"
| [1] | TILMAN D, BALZER C, HILL J, BEFORT B L. Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences of the United States of America, 2011,108(50):20260-20264. |
| [2] | BARTON L, NEWSOME S D, CHEN F H, WANG H, GUILDERSON T P, BETTINGER R L. Agricultural origins and the isotopic identity of domestication in northern China. Proceedings of the National Academy of Sciences of the United States of America, 2009,106(14):5523-5528. |
| [3] |
JIA G Q, HUANG X H, ZHI H, ZHAO Y, ZHAO Q, LI W J, CHAI Y, YANG L, LIU K Y, LU H Y, ZHU C R, LU Y Q, ZHOU C C, FAN D L, WENG Q J, GUO Y L, HUANG T, ZHANG L, LU T T, FENG Q, HAO H F, LIU H K, LU P, ZHANG N, LI Y H, GUO E, WANG S J, WANG S Y, LIU J R, ZHANG W F, CHEN G Q, ZHANG B G, LI W, WANG Y F, LI H Q, ZHAO B H, LI J Y, DIAO X M, HAN B. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nature Genetics, 2013,45(8):957-961.
doi: 10.1038/ng.2673 |
| [4] |
BENNETZEN J L, SCHMUTZ J, WANG H, PERCIFIELD R, HAWKINS J, PONTAROLI A C, ESTEP M, FENG L, VAUGHN J N, GRIMWOOD J, JENKINS J, BARRY K, LINDQUIST E, HELLSTEN U, DESHPANDE S, WANG X W, WU X M, MITROS T, TRIPLETT J, YANG X H, YE C Y, MAURO-HERRERA M, WANG L, LI P H, SHARMA M, SHARMA R, RONALD P C, PANAUD O, KELLOGG E A, BRUTNELL T P, DOUST A N, TUSKAN G A, ROKHSAR D, DEVOS K M. Reference genome sequence of the model plant Setaria. Nature Biotechnology, 2012,30(6):555-561.
doi: 10.1038/nbt.2196 |
| [5] |
ZHANG G Y, LIU X, QUAN Z W, CHENG S F, XU X, PAN S K, XIE M, ZENG P, YUE Z, WANG W L, TAO Y, BIAN C, HAN C L, XIA Q J, PENG X H, CAO R, YANG X H, ZHAN D L, HU J C, ZHANG Y X, LI H N, LI H, LI N, WANG J Y, WANG C C, WANG R Y, GUO T, CAI Y J, LIU C Z, XIANG H T, SHI Q X, HUANG P, CHEN Q C, LI Y R, WANG J, ZHAO Z H, WANG J. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature Biotechnology, 2012,30(6):549-554.
doi: 10.1038/nbt.2195 |
| [6] |
YANG Z R, ZHANG H S, LI X K, SHEN H M, GAO J H, HOU S Y, ZHANG B, MAYES S, BENNETT M, MA J X, WU C Y, SUI Y, HAN Y H, WANG X C. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nature Plants, 2020,6(9):1167-1178.
doi: 10.1038/s41477-020-0747-7 |
| [7] |
COLASANTI J, CONEVA V. Mechanisms of floral induction in grasses: Something borrowed, something new. Plant Physiology, 2009,149(1):56-62.
doi: 10.1104/pp.108.130500 |
| [8] |
ANDRÉS F, COUPLAND G. The genetic basis of flowering responses to seasonal cues. Nature Review Genetics, 2012,13(9):627-639.
doi: 10.1038/nrg3291 |
| [9] |
RIESEBERG L H, WILLIS J H. Plant speciation. Science, 2007,317(5840):910-914.
doi: 10.1126/science.1137729 |
| [10] |
STRAYER C, OYAMA T, SCHULTZ T F, RAMAN R, SOMERS D E, MÁS P, PANDA S, KREPS J A, KAY S A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science, 2000,289(5480):768-771.
doi: 10.1126/science.289.5480.768 |
| [11] |
COCKRAM J, THIEL T, STEUERNAGEL B, STEIN N, TAUDIEN S, BAILEY P C, O'SULLIVAN D M. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS ONE, 2012,7(9):e45307.
doi: 10.1371/journal.pone.0045307 |
| [12] |
WENKEL S, TURCK F, SINGER K, GISSOT L, LE GOURRIEREC J L, SAMACH A, COUPLAND G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell, 2006,18(11):2971-2984.
doi: 10.1105/tpc.106.043299 |
| [13] | GENDRON J M, PRUNEDA-PAZ J L, DOHERTY C J, GROSS A M, KANG S E, KAY S A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proceedings of the National Academy of Sciences of the United States of America, 2012,109(8):3167-3172. |
| [14] |
MAKINO S, MATSUSHIKA A, KOJIMA M, YAMASHINO T, MIZUNO T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1- overexpressing plants. Plant Cell Physiology, 2002,43(1):58-69.
doi: 10.1093/pcp/pcf005 |
| [15] |
SHIM J S, KUBOTA A, IMAIZUMI T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiology, 2017,173(1):5-15.
doi: 10.1104/pp.16.01327 |
| [16] |
KOO B H, YOO S C, PARK J W, KWON C T, LEE B D, AN G, ZHANG Z, LI J, LI Z, PAEK N C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Molecular Plant, 2013,6(6):1877-1888.
doi: 10.1093/mp/sst088 |
| [17] |
SALOMÉP A, MCCLUNG C R. The Arabidopsis thaliana clock. Journal of Biological Rhythms, 2004,19(5):425-435.
doi: 10.1177/0748730404268112 |
| [18] |
MÁS P. Circadian clock signaling in Arabidopsis thaliana: From gene expression to physiology and development. The International Journal of Developmental Biology, 2005,49(5/6):491-500.
doi: 10.1387/ijdb.041968pm |
| [19] |
GARDNER M J, HUBBARD K E, HOTTA C T, DODD A N, WEBB A A. How plants tell the time. Biochemical Journal, 2006,397(1):15-24.
doi: 10.1042/BJ20060484 |
| [20] |
ALABADÍ D, OYAMA T, YANOVSKY M J, HARMON F G, MÁS P, KAY S A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science, 2001,293(5531):880-883.
doi: 10.1126/science.1061320 |
| [21] |
PRUNEDA-PAZ J L, BRETON G, PARA A, KAY S A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science, 2009,323(5920):1481-1485.
doi: 10.1126/science.1167206 |
| [22] | 陆平. 谷子种质资源描述规范和数据标准2-9. 北京: 中国农业出版社, 2006. |
| LU P. Description Specification and Data Standard of Foxtail Millet Germplasm Resources 2-9. Beijing: China Agriculture Press, 2006. (in Chinese) | |
| [23] | TURNER S D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv, 2014, https://doi.org/10.1101/005165. |
| [24] |
YANG A, DAI X Y, ZHANG W H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. Journal of Experimental Botany, 2012,63(7):2541-2556.
doi: 10.1093/jxb/err431 |
| [25] |
LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 2001,25(4):402-408.
doi: 10.1006/meth.2001.1262 |
| [26] | DOYLE J. DNA protocols for plants-CTAB total DNA isolation. Molecular Techniques in Taxonomy, 1991: 283-293. |
| [27] |
LIBRADO P, ROZAS J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 2009,25(11):1451-1452.
doi: 10.1093/bioinformatics/btp187 |
| [28] | 刁现民, 程汝宏. 十五年区试数据分析展示谷子糜子育种现状. 中国农业科学, 2017,50(23):4469-4474. |
| DIAO X M, CHENG R H. Fifteen-year regional trial data analysis shows the current situation of millet and millet breeding. Scientia Agricultura Sinica, 2017,50(23):4469-4474. (in Chinese) | |
| [29] |
YANO M, KATAYOSE Y, ASHIKARI M, YAMANOUCHI U, MONNA L, FUSE T, BABA T, YAMAMOTO K, UMEHARA Y, NAGAMURA Y, SASAKI T. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell, 2000,12(12):2473-2484.
doi: 10.1105/tpc.12.12.2473 |
| [30] |
HAYAMA R, YOKOI S, TAMAKI S, YANO M, SHIMAMOTO K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature, 2003,422(6933):719-722.
doi: 10.1038/nature01549 |
| [31] |
XUE W Y, XING Y Z, WENG X Y, ZHAO Y, TANG W J, WANG L, ZHOU H J, YU S B, XU C G, LI X H, ZHANG Q F. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics, 2008,40(6):761-767.
doi: 10.1038/ng.143 |
| [32] |
NEMOTO Y, NONOUE Y, YANO M, IZAWA T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. The Plant Journal, 2016,86(3):221-233.
doi: 10.1111/tpj.2016.86.issue-3 |
| [33] |
DU A, TIAN W, WEI M H, YAN W, HE H, ZHOU D, HUANG X, LI S G, OUYANG X H. The DTH8-Hd1 module mediates day-length- dependent regulation of rice flowering. Molecular Plant, 2017,10(7):948-961.
doi: 10.1016/j.molp.2017.05.006 |
| [34] |
FUJINO K, YAMANOUCHI U, NONOUE Y, OBARA M, YANO M. Switching genetic effects of the flowering time gene Hd1 in LD conditions by Ghd7 and OsPRR37 in rice. Breeding Science, 2019,69(1):127-132.
doi: 10.1270/jsbbs.18060 |
| [35] |
ZHANG Z Y, ZHANG B, QI F X, WU H, LI Z X, XING Y Z. Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice. Molecular Breeding, 2019,39(92):1-12.
doi: 10.1007/s11032-018-0907-x |
| [1] | 李周帅,董远,李婷,冯志前,段迎新,杨明羡,徐淑兔,张兴华,薛吉全. 基于杂交种群体的玉米产量及其配合力的全基因组关联分析[J]. 中国农业科学, 2022, 55(9): 1695-1709. |
| [2] | 职蕾,者理,孙楠楠,杨阳,Dauren Serikbay,贾汉忠,胡银岗,陈亮. 小麦苗期铅耐受性的全基因组关联分析[J]. 中国农业科学, 2022, 55(6): 1064-1081. |
| [3] | 贾冠清, 刁现民. 中国谷子种业创新现状与未来展望[J]. 中国农业科学, 2022, 55(4): 653-665. |
| [4] | 黄勋和,翁茁先,李威娜,王庆,何丹林,罗威,张细权,杜炳旺. 中国地方品种黄鸡线粒体DNA D-loop遗传多样性研究[J]. 中国农业科学, 2022, 55(22): 4526-4538. |
| [5] | 逄洪波, 程露, 于茗兰, 陈强, 李玥莹, 吴隆坤, 王泽, 潘孝武, 郑晓明. 栽培稻芽期耐低温全基因组关联分析[J]. 中国农业科学, 2022, 55(21): 4091-4103. |
| [6] | 谢晓宇, 王凯鸿, 秦晓晓, 王彩香, 史春辉, 宁新柱, 杨永林, 秦江鸿, 李朝周, 马麒, 宿俊吉. 陆地棉吐絮率的限制性两阶段多位点全基因组关联分析及候选基因预测[J]. 中国农业科学, 2022, 55(2): 248-264. |
| [7] | 邹林翰,周新颖,张泽源,蔚睿,袁梦,宋晓朋,简俊涛,张传量,韩德俊,宋全昊. 小麦周8425B×小偃81重组自交系群体千粒重相关性状的QTL定位及单倍型分析[J]. 中国农业科学, 2022, 55(18): 3473-3483. |
| [8] | 常立国,何坤辉,刘建超. 多环境下玉米保绿相关性状遗传位点的挖掘[J]. 中国农业科学, 2022, 55(16): 3071-3081. |
| [9] | 郭淑青,宋慧,柴少华,郭岩,石兴,杜丽红,邢璐,解慧芳,张扬,李龙,冯佰利,刘金荣,杨璞. 谷子生育期及穗相关性状的QTL定位[J]. 中国农业科学, 2022, 55(15): 2883-2898. |
| [10] | 李婷,董远,张君,冯志前,王亚鹏,郝引川,张兴华,薛吉全,徐淑兔. 玉米杂交种穗部性状的全基因组关联分析[J]. 中国农业科学, 2022, 55(13): 2485-2499. |
| [11] | 王娟, 马晓梅, 周小凤, 王新, 田琴, 李成奇, 董承光. 棉花产量构成因素性状的全基因组关联分析[J]. 中国农业科学, 2022, 55(12): 2265-2277. |
| [12] | 梁鹏,张天闻,孟科,邵顺成,邹诗凡,荣轩,强浩,冯登侦. 绵羊ADIPOQ多态性与生长性状的关联分析[J]. 中国农业科学, 2022, 55(11): 2239-2256. |
| [13] | 崔承齐, 刘艳阳, 江晓林, 孙知雨, 杜振伟, 武轲, 梅鸿献, 郑永战. 芝麻产量相关性状的多位点全基因组关联分析及候选基因预测[J]. 中国农业科学, 2022, 55(1): 219-232. |
| [14] | 马拴红, 万炯, 梁瑞清, 张雪海, 邱小倩, 孟淑君, 徐宁坤, 林源, 党昆泰, 王琪月, 赵嘉雯, 丁冬, 汤继华. 玉米开花期转录因子候选基因的关联分析[J]. 中国农业科学, 2022, 55(1): 12-25. |
| [15] | 武翠卿,孙静鑫,郭平毅,王宏富,武新慧. 农艺措施对谷子产量及抗倒伏力学性能的影响[J]. 中国农业科学, 2021, 54(6): 1127-1142. |
|
||