













中国农业科学 ›› 2019, Vol. 52 ›› Issue (12): 2171-2182.doi: 10.3864/j.issn.0578-1752.2019.12.014
收稿日期:2018-07-15
接受日期:2019-04-15
出版日期:2019-06-16
发布日期:2019-06-22
通讯作者:
刘远,黄勤楼
作者简介:李文杨,Tel:0591-87902045;E-mail: 516316606@qq.com。
基金资助:
LI WenYang,LIU Yuan( ),WU XianFeng,GAO ChengFang,HUANG QinLou(
),WU XianFeng,GAO ChengFang,HUANG QinLou( )
)
Received:2018-07-15
Accepted:2019-04-15
Online:2019-06-16
Published:2019-06-22
Contact:
Yuan LIU,QinLou HUANG
摘要:
【目的】 研究福清山羊与努比亚黑山羊发情期卵巢组织转录组差异表达水平,一方面为山羊繁殖性状形成的分子机制提供理论依据,另一方面为利用努比亚黑山羊杂交改良福清山羊提供可加快遗传进展的分子标记。【方法】 利用转录组测序方法对福清山羊和努比亚黑山羊发情期卵巢组织进行研究,筛选品种间的差异表达基因(differentially expressed genes,DEGs),并对DEGs功能进行注释和若干基因荧光定量PCR(quantitative real-time PCR,qRT-PCR)验证;同时通过与参考基因组比对,分析和筛选测序样品中存在的SNP/InDel。【结果】 6个样品共得到46.68Gb Clean Data,DESeq分析发现了福清山羊和努比亚黑山羊发情期卵巢组织的DEGs149个(包含25个新转录本),其中表达上调53个,表达下调96个;初步认为输卵管素基因(oviductin,OVN)、类固醇合成快速调节蛋白基因(steroidogenic acute regulatory protein,STAR)、早期生长应答1基因(early growth response 1,EGR1)可作为福清山羊繁殖性能的候选基因。149个DEGs中的108个基因能被GO(gene ontology)数据库注释,30个DEGs能够被COG(Cluster of Orthologous Groups of proteins)数据库注释,91个DEGs能够被KEGG(kyoto encyclopediaof genes and genomes)数据库注释。KEGG通路分析表明DEGs共富集到21条信号通路中,6条通路显著富集。经过进一步的与参考基因组序列比对分析,共发掘1 506个新转录本。经qRT-PCR验证,所选基因(转录本)表达变化模式与转录组测序结果一致,表明测序结果可靠。【结论】 在转录组水平上筛选出了福清山羊和努比亚黑山羊发情期卵巢组织的149个DEGs,发掘了1 506个新转录本,初步揭示了OVN、STAR和EGR1在山羊繁殖过程中可能发挥重要作用,为进一步探索山羊繁殖性状相关机理提供参考依据。
李文杨,刘远,吴贤锋,高承芳,黄勤楼. 福清山羊与努比亚黑山羊发情期卵巢组织RNA-Seq分析[J]. 中国农业科学, 2019, 52(12): 2171-2182.
LI WenYang,LIU Yuan,WU XianFeng,GAO ChengFang,HUANG QinLou. Transcriptome Analysis of Differentially Gene Expression Associated with Ovary Tissue During the Follicular Stage in Fuqing Goat and Nubian Black Goat[J]. Scientia Agricultura Sinica, 2019, 52(12): 2171-2182.
表1
实时荧光定量PCR引物信息"
| 基因名称 Gene | 上游引物 Forward primer(5′-3′) | 下游引物 Reverse primer(5′-3′) | 产物长度 Product size(bp) |
|---|---|---|---|
| 18S rRNA | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG | 151 |
| New-Gene 4708 | CTCCAAATCGGGCAACACAG | TCCACAGTAATAATCGGCCTCG | 77 |
| New-Gene 1125 | ACAACAGTGTCAGCATCCGT | TGTTGGAGTCCCGTTATGGC | 136 |
| CREB3L4 | GGGCTCTTCGAAACAGCAGA | AGTCCCAGCTCCAGGAAAGA | 104 |
| THSD7B | AGTTACAAGGCGCATCACCA | TTTCGGCATGATGCAGGACT | 159 |
| CRABP1 | GTGCAGGAGCTTACCCACTT | CGGGTCCAGTAGGTTTTGGG | 93 |
| CTHRC1 | TTGGTGCTGGACTAGTGGAC | ATGCGAGACACCGAATTCCA | 100 |
表2
测序数据与参考基因组的序列比对结果统计"
| 样品 Sample | 总Reads Total reads | 可定位的Reads Mapped reads | 唯一定位Reads Uniq mapped reads | 多点定位Reads Multiple mapped reads |
|---|---|---|---|---|
| F-1 | 48567844 | 41489885/85.43% | 39687502/81.72% | 1802383/3.71% |
| F-2 | 49910620 | 42262711/84.68% | 40565566/81.28% | 1697145/3.40% |
| F-3 | 49890394 | 42216928/84.62% | 40925490/82.03% | 1791438/3.59% |
| N-4 | 53482074 | 46603840/87.14% | 45110457/84.35% | 1293383/2.42% |
| N-5 | 53379622 | 46077636/86.32% | 44696951/83.73% | 1380685/2.59% |
| N-6 | 53967784 | 46261607/85.72% | 45017922/83.42% | 1243685/2.30% |
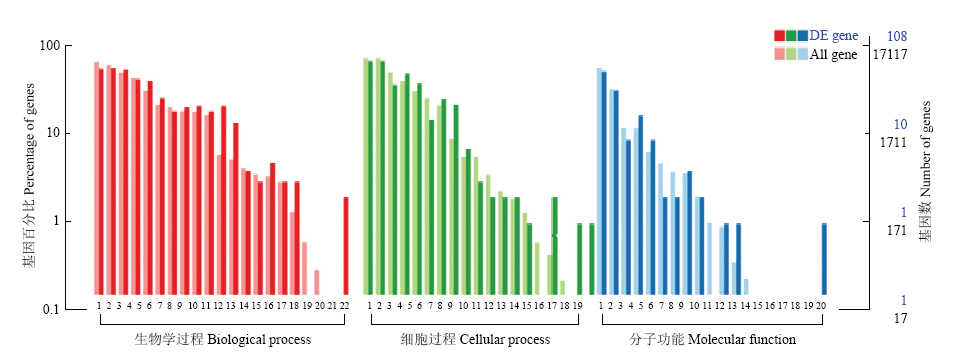
图3
差异表达基因GO注释 生物学过程Biological process:1细胞过程 Cellular process;2单一的生物过程 Single-organism process;3生物调节 Biological regulation;4代谢过程 Metabolic process;5刺激反应 Response to stimulus;6多细胞生物过程 Multicellular organismal process;7信号 Signaling;8定位 Localization;9发育过程 Developmental process;10组织或生物起源细胞组件 Cellular component organization or biogenesis;11免疫系统过程 Immune system process;12多生物体过程 Multi-organism process;13繁殖过程Reproductive process;14移动Locomotion;15生物粘附Biological adhesion;16繁殖Reproductive;17生长Growth;18有节奏的过程Rhythmic process;19激素分泌 Hormone secretion;20生物相Biological phase;21细胞杀伤 Cell killing;22细胞聚集Cell aggregation"
| [1] | 薛慧良, 周忠孝 . 绵羊繁殖性状候选基因的研究进展. 动物科学与动物医学, 2003,20(3):32-35. |
| XUE H L, ZHOU X Z . Advances in candidate genes for reproductive traits in sheep. Animal Science and Veterinary Medicine, 2003, 20(3):32-35. (in Chinese) | |
| [2] | 刘远, 林碧芬, 林仕欣, 李文杨, 张晓佩, 高承芳, 董晓宁 . 浅谈福建地方山羊品种高繁殖力资源的开发利用. 中国草食动物科学, 2010,30(6):57-59. |
| LIU Y, LIN B F, LIN S X, LI W Y, ZHANG X P, GAO C F, DONG X N . Exploitation and utilization of high fertility resources of Fujian local goat breeds. China Herbivore Science, 2010,30(6):57-59. (in Chinese) | |
| [3] | LING Y H, XIANG H, LI Y S, LIU Y, ZHANG Y H, ZHANG Z J, DING J P, ZHANG X R . Exploring differentially expressed genes in the ovaries of uniparous and multiparous goats using the RNA-Seq (quantification) method. Gene, 2004,550(1):148-153. |
| [4] | VALERIO C, CLAUDIA A, ITALIA D F, ALFREDO C . Uncovering the complexity of transcriptomes with RNA-Seq. Journal of Biomedicine and Biotechnology, 2010,2010:1-19. |
| [5] |
WANG Z, GERSTEIN M, SNYDER M . RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics, 2009,10(1):57-63.
doi: 10.1038/nrg2484 |
| [6] | 李丽, 缪中伟, 辛清武, 朱志明, 章琳俐, 庄晓东, 郑嫩珠 . 半番鸭与番鸭精巢组织差异表达转录组测序分析. 中国农业科学, 2017,50(18):3608-3619. |
| LI L, MIAO Z W, XIN Q W, ZHU ZM, ZHANG L L, ZHUANG X D, ZHENG N Z . Transcriptome analysis of differentially gene expression associated with testis tissue in mule duck and Muscovy duck. Scientia Agricultura Sinica, 2017,50(18):3608-3619. (in Chinese) | |
| [7] | 字向东, 罗斌, 夏威, 郑玉才, 熊显荣, 李键, 钟金城, 朱江江, 张正帆 . 基于RNA-Seq技术的牦牛体外受精胚胎发育转录组分析. 中国农业科学, 2018,51(8):1577-1589. |
| ZI X D, LUO B, XIA W, ZHENG Y C, XIONG X R, LI J, ZHONG J C, ZHU J J, ZHANG Z F . Transcripomic analysis of IVF embryonic development in the yak( Bos grunniens) Via RNA-Seq. Scientia Agricultura Sinica, 2018, 51(8):1577-1589. (in Chinese) | |
| [8] | 罗庆苗, 秦晓玉, 苗向阳 . 利用数字基因表达谱对小尾寒羊高繁性状相关基因的筛选研究. 中国农学通报, 2012,28(23):18-25. |
| LUO Q M, QIN X Y, MIAO X Y . The study on related genes of high fecundity using digital gene expression tag profiling in small-tail- han-sheep. Chinese Agricultural Science Bulletin, 2012, 28(23):18-25. (in Chinese) | |
| [9] | DI R, HE J N, SONG S H, TIAN D M, LIU Q Y, LIANG X J, MA Q, SUN M, WANG J D, ZGAO W M, CAO G L, WANG J X, YANG Z M, GE Y, CHU M X . Characterization and comparative profiling of ovarian microRNAs during ovine anestrus and the breeding season. BMC Genomics, 2014, 15:899. |
| [10] | FENG X, KI F Z, WANG F, ZHANG G M, PANG J, REN C F, ZHANG T T, YANG H, WANG Z Y, ZHANG Y L . Genome-wide differential expression profiling of mRNAs and IncRNAs associated with prolificacy in Hu sheep. Bioscience Reports, 2018,38, 1-14. |
| [11] | 付绍印, 何小龙, 王标, 李虎山, 储明星, 刘永斌, 张文广 . 不同产羔数绵羊性腺轴比较转录组研究. 中国畜牧兽医, 2017,44(2):488-496. |
| FU S Y, HE X L, WANG B, LI H S, CHU M X, LIU Y B, ZHANG W G . Comparative Transcriptome Research of Gonadal Axis for Different Lambing Number of Sheep. China Animal Husbandry and Veterinary Medicine, 2017, 44(2):488-496. (in Chinese) | |
| [12] | 李文杨, 刘远, 林仕欣, 张晓佩, 高承芳, 林碧芬, 董晓宁 . BMPR-IB, BMP15, GDF9基因作为福建省4个主要山羊品种多胎性能候选基因的研究. 浙江农业学报, 2012,24(6):975-979. |
| LI W Y, LIU Y, LIN S X, ZHANG X P, GAO C F, LIN B F, DONG X N . Studies on BMPR-IB, BMP15, and GDF9 as candidate genes of fecundity in four breeds of goats in Fujian. Acta Agriculture Zhejiang, 2012,24(6):975-979. (in Chinese) | |
| [13] | 陈其新, 张建红, 宋彦军, 董济福 . 我国主要肉羊品种肉用性能的初步评价. 中国草食动物科学(全国养羊生产与学术研讨会议论文集), 2012(s1):357-362. |
| CHEN Q X, ZHANG J H, SONG Y J, DONG J F . Preliminary evaluation of meat performance of main mutton sheep and goats in China.China Herbivore Science, 2012(s1):357-362. (in Chinese) | |
| [14] | 洪开祥, 余荣权, 徐科礼, 陈太金, 许永强, 王清颖 . 努比亚羊与贵州黑山羊杂交性能研究. 中国草食动物科学(全国养羊生产与学术研讨会议论文集), 2014(s1):154-156. |
| HONG X K, YU R Q, XU K L, CHEN T J, XU Y Q, WANG Q Y . Study on hybrid performance of Nubian goat and Guizhou black goat.China Herbivore Science, 2012(s1):357-362. (in Chinese) | |
| [15] |
KIM D, PERTEA G, TRAPNELL C, PIMENTEL H, KELLEY R, SALZBERG S L . TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 2013,14:R36.
doi: 10.1186/gb-2013-14-4-r36 |
| [16] | DONG Y, XIE M, JIANG Y, XIAO N Q, DU X Y, ZHANG W G, TOSSER K G, WANG J H, YANG S, LIANG J, CHEN W B, CHEN J, ZENG P, HOU Y, BIAN C, PAN S K, LI Y X, LIU X, WANG W L, SERVIN B, SAYRE B, ZHU B, SWEENEY D, MOORE R, NIE W H, SHEN Y Y, ZHAO R P, ZHANG G J, LI J Q, FARAUT T, WOMACK J, ZHANG Y P, KIJAS J, COCKETT N, XU X, ZHANG S H, WANG J, WANG W . Sequencing and automated whole-genome optical mapping of the genome of a domestic goat ( Capra hircus). Nature Biotechnology, 2013,31(2):135-141. |
| [17] |
JIANG H, WONG W H . Statistical inferences for isoform expression in RNA-Seq. Bioinformatics, 2009,25(8):1026-1032.
doi: 10.1093/bioinformatics/btp113 |
| [18] |
FLOREA L, SONG L, SALZBERG S L . Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000 Research, 2013,2:188.
doi: 10.12688/f1000research |
| [19] |
SCHULZE S K, KANWAR R, GOLZENLEUCHTER M, THRNEAU T M, BEYTLER A S . SERE: Single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics, 2012,13(1):524.
doi: 10.1186/1471-2164-13-524 |
| [20] |
WANG L K, FENG Z X, WANG X, WANG X W, ZHANG X G . DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics, 2010,26, 136-138.
doi: 10.1093/bioinformatics/btp612 |
| [21] | ALEXA A, RAHNENFUHRER J . TopGO: enrichment analysis for gene ontology. R package version 2. 8, 2010. |
| [22] | TATUSOV R L, GALPERIN M Y, NATALE D A . The COG database: a tool for genome scale analysis of protein functions and evolution. Nucleic Acids Research, 2000, 28(1):33-36. |
| [23] |
KANEHISA M, GOTO S, KAWASHIMA S, OKUNO Y, HATTORI M . The KEGG resource for deciphering the genome. Nucleic Acids Research, 2004,32:277-280.
doi: 10.1093/nar/gkh063 |
| [24] |
MCKENNA A, HANNA M, BANKS E, SIVACHENKO A, CIBULSKIS K, KERNYTSKY A, GARIMELLA K, ALTSHULER D, GABRIEL S, DALY M, DEPRISTO M A . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 2010,20(9):1297-1303.
doi: 10.1101/gr.107524.110 |
| [25] | CINGOLANI P, PLATTS A, WANG L L, COON M, NGUYEN T, WANG L, LAND S J, LU X, RUDEN D M . A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012,6(2):80-92. |
| [26] | 梁素芸, 周正奎, 侯水生 . 基于测序技术的畜禽基因组学研究进展. 遗传, 2017,39(4):276-292. |
| LIANG S Y, ZHOU Z K, HOU S S . The research progress of farm animal genomics based on sequencing technologies. Hereditas (Beijing), 2017, 39(4):276-292. (in Chinese) | |
| [27] | 兰道亮, 熊显荣, 位艳丽, 徐通, 钟金城, 字向东, 王永, 李健 . 基于RNA-Seq高通量测序技术得牦牛卵巢转录组研究: 进一步完善牦牛基因结构及挖掘与繁殖相关新基因. 中国科学(生命科学), 2014,44(3):307-317. |
| LAN D L, XIONNG X R, WEI Y L, XU T, ZHONG J C, ZI X D, WANG Y, LI J . RNA-Seq analysis of yak ovary: improving yak gene structure information and mining reproduction-related genes. Science China-Life Sciences , 2014,44(3):307-317. (in Chinese) | |
| [28] | 王旭平, 杨胜林, 陆曼, 柳诚刚, 谭斌, 杨汝才, 周美迪, 王达娜, 李潇蒙 . 番鸭攻击行为的转录组学研究. 农业生物技术学报, 2018,26(4):687-697. |
| WANG X P, YANG S L, LU M, LIU C G, TAN B, YANG R C, ZHOU M D, WANG D N, LI X M . Transcriptomics analysis of the aggressive behavior of Muscovy Duck ( Cairna moschata). Journal of Agricultural Biotechnology, 2018,26(4):687-697. (in Chinese) | |
| [29] |
BUHI W C . Characterization and biological roles of oviduct-specific, oestrogen-dependent glycoprotein. Reproduction, 2002,123(3):355-362.
doi: 10.1530/rep.0.1230355 |
| [30] | 张译夫, 潘阳阳, 崔燕, 余四九, 张倩, 樊江峰, 杨琨, 杨融 . 牦牛输卵管蛋白(Ovn)的cDNA克隆及其在发情周期中的差异性表达. 农业生物技术学报, 2015,23(9):1217-1225. |
| ZHANG Y F, PAN Y Y, CUI Y, YU S J, ZHANG Q, PAN J F, YANG K , YANG R. cDNA cloning of Yak ( Bos grunniens) Oviductin (Ovn) and its different expression during the estrous cycle. Journal of Agricultural Biotechnology, 2015,23(9):1217-1225. (in Chinese) | |
| [31] | VERHAGE H G, MAVROGIANIS P A , O'DAY B, SCHMIDT A, RIAS E B, DONNELLY K M, BOOMSMA R A, THIBODEAUX J K, FAZLEABAS A T, JAFFE R C. Characteristics of an oviductal glycoprotein and its potential role in the fertilization process. Biology of Reproduction, 1998, 58(5):1098-1101. |
| [32] | SKIBOLA C F, BRACCI P M, HALPERIN E, NIETERS A, HUBBARD A, PAYNTER R A, SKIBOLA D R, AGANA L, BECKER N, TRESSLER P, FORREST M . Polymorphisms in the estrogen receptor 1 and Vitamin C and Matrix metalloproteinase gene families are associated with susceptibility to lymphoma. PLoS One, 2008,7(3):e2816. |
| [33] | SAYASITH K, BROWN K A, LUSSIER J G, DORÉ M, SIROIS J . Characterization of bovine early growth response factor-1 and its gonadotropin-dependent regulation in ovarian follicles prior to ovulation. Journal of Molecular Endocrinology, 2006, 37:239-250. |
| [34] | 吴阳升, 林嘉鹏, 汪立芹, 陈莹, 黄俊成 . 绵羊小卵泡与中卵泡转录组差异特征分析. 江苏农业学报, 2016,32(4):832-843. |
| WU Y S, LIN J P, WANG L Q, CHEN Y, HUANG J C . Transcriptome profiling of ovine follicles during growth from small to middle antral sizes. Jiangsu Journal of Agriculture Science, 2016,32(4):832-843. (in Chinese) | |
| [35] |
MIAO X, QIN Q L . Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity. Molecular and Cellular Endocrinology, 2015,402(C):32.
doi: 10.1016/j.mce.2014.12.023 |
| [36] |
段新崇, 魏彦辉, 李阳, 李相运, 周荣艳, 锡建中 . 小尾寒羊间情期和发情期microRNAs差异表达分析. 畜牧兽医学报, 2016,47(7):1324-1332.
doi: 10.11843/j.issn.0366-6964.2016.07.004 |
|
DUAN X C, WEI Y H, LI Y, LI X Y, ZHOU R Y, XI J Z . Analyzing the differential expression of microRNAs in estrus and diestrus of small tailed Han sheep. Acta Veterinaria et Zootechnica Sinica, 2016,47(7):1324-1332. (in Chinese)
doi: 10.11843/j.issn.0366-6964.2016.07.004 |
| [1] | 刘玉芳,陈玉林,周祖阳,储明星. miR-221-3p靶向BCL2L11调控小尾寒羊卵泡颗粒细胞凋亡[J]. 中国农业科学, 2022, 55(9): 1868-1876. |
| [2] | 阿依木古丽·阿不都热依木,阿尔祖古丽·阿依丁,王家敏,石嘉琛,马芳芳,蔡勇,乔自林. 大豆异黄酮对牦牛卵巢颗粒细胞增殖和凋亡的影响[J]. 中国农业科学, 2022, 55(8): 1667-1675. |
| [3] | 由玉婉,张雨,孙嘉毅,张蔚. ‘月月粉’月季NAC家族全基因组鉴定及皮刺发育相关成员的筛选[J]. 中国农业科学, 2022, 55(24): 4895-4911. |
| [4] | 尤佳玲,李有梅,孙孟豪,谢兆森. ‘黑比诺’葡萄不同叶龄叶片叶绿体内淀粉积累及其相关基因表达差异分析[J]. 中国农业科学, 2022, 55(21): 4265-4278. |
| [5] | 孙保娟,汪瑞,孙光闻,王益奎,李涛,宫超,衡周,游倩,李植良. 转录组及代谢组联合解析茄子果色上位遗传效应[J]. 中国农业科学, 2022, 55(20): 3997-4010. |
| [6] | 刘鑫,张亚红,袁苗,党仕卓,周娟. ‘红地球’葡萄花芽分化过程中的转录组分析[J]. 中国农业科学, 2022, 55(20): 4020-4035. |
| [7] | 李丽莹,何颖婷,钟玉宜,周小枫,张豪,袁晓龙,李加琪,陈赞谋. CTNNB1对猪卵巢颗粒细胞的调控[J]. 中国农业科学, 2022, 55(15): 3050-3061. |
| [8] | 张静,张姬越,岳永起,赵丹,范依琳,马妍,熊燕,熊显荣,字向东,李键,杨丽雪. LKB1基因对卵巢颗粒细胞类固醇激素生成相关基因的调控作用[J]. 中国农业科学, 2022, 55(10): 2057-2066. |
| [9] | 徐献斌,耿晓月,李慧,孙丽娟,郑焕,陶建敏. 基于转录组分析ABA促进葡萄花青苷积累相关基因[J]. 中国农业科学, 2022, 55(1): 134-151. |
| [10] | 郭永春, 王鹏杰, 金珊, 侯炳豪, 王淑燕, 赵峰, 叶乃兴. 基于WGCNA鉴定茶树响应草甘膦相关的基因共表达模块[J]. 中国农业科学, 2022, 55(1): 152-166. |
| [11] | 陈华枝,范元婵,蒋海宾,王杰,范小雪,祝智威,隆琦,蔡宗兵,郑燕珍,付中民,徐国钧,陈大福,郭睿. 基于纳米孔全长转录组数据完善东方蜜蜂微孢子虫的基因组注释[J]. 中国农业科学, 2021, 54(6): 1288-1300. |
| [12] | 杜宇,祝智威,王杰,王秀娜,蒋海宾,范元婵,范小雪,陈华枝,隆琦,蔡宗兵,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 利用第三代纳米孔长读段测序技术构建和注释蜜蜂球囊菌的全长转录组[J]. 中国农业科学, 2021, 54(4): 864-876. |
| [13] | 赵卫松,郭庆港,董丽红,王培培,苏振贺,张晓云,鹿秀云,李社增,马平. 枯草芽孢杆菌NCD-2对棉花根系分泌物L-脯氨酸响应的转录-蛋白质组学联合分析[J]. 中国农业科学, 2021, 54(21): 4585-4600. |
| [14] | 刘恋,唐志鹏,李菲菲,熊江,吕壁纹,马小川,唐超兰,李泽航,周铁,盛玲,卢晓鹏. ‘融安金柑’‘滑皮金柑’及‘脆蜜金柑’贮藏期品质、贮藏特性及果皮转录组分析[J]. 中国农业科学, 2021, 54(20): 4421-4433. |
| [15] | 杜星,曾强,刘禄,李琦琦,杨柳,潘增祥,李齐发. 二花脸猪linc-NORFA核心启动子鉴定与转录调控分析[J]. 中国农业科学, 2021, 54(15): 3331-3342. |
|
||