













中国农业科学 ›› 2021, Vol. 54 ›› Issue (22): 4709-4727.doi: 10.3864/j.issn.0578-1752.2021.22.001
吕士凯( ),马小龙,张敏,邓平川,陈春环,张宏(
),马小龙,张敏,邓平川,陈春环,张宏( ),刘新伦(
),刘新伦( ),吉万全(
),吉万全( )
)
收稿日期:2021-02-10
接受日期:2021-04-07
出版日期:2021-11-16
发布日期:2021-11-19
通讯作者:
张宏,刘新伦,吉万全
作者简介:吕士凯,E-mail: 基金资助:
LÜ ShiKai( ),MA XiaoLong,ZHANG Min,DENG PingChuan,CHEN ChunHuan,ZHANG Hong(
),MA XiaoLong,ZHANG Min,DENG PingChuan,CHEN ChunHuan,ZHANG Hong( ),LIU XinLun(
),LIU XinLun( ),JI WanQuan(
),JI WanQuan( )
)
Received:2021-02-10
Accepted:2021-04-07
Online:2021-11-16
Published:2021-11-19
Contact:
Hong ZHANG,XinLun LIU,WanQuan JI
摘要:
【目的】以条锈菌和白粉菌胁迫的普通小麦(Triticum aestivum L.)为研究对象,分析由可变剪切(alternative splicing,AS)形成的TaNAC结构变异转录本,同时分析TaNAC基因的microRNA调控位点,为进一步解析TaNAC基因通过转录后调控参与小麦响应真菌胁迫奠定基础。【方法】普通小麦兼抗种质N9134在被白粉菌和条锈菌分别侵染后,各8个时间点取样并混合,然后从混合样本中克隆得到大量TaNAC转录本。参考中国春小麦基因组注释信息(IWGSC RefSeq v1.1)进行比对,选择由可变剪切形成的TaNAC序列结构变异转录本,分析它们的序列结构特征。利用生物信息学软件和在线工具,对这些TaNAC结构变异转录本编码产物的功能结构域、高级结构、理化性质、亚细胞定位等特征和变异情况进行比对分析。同时,利用洋葱表皮细胞瞬时表达系统验证其中1对TaNAC结构变异转录本的亚细胞定位预测结果,并选取5组TaNAC基因的可变剪切序列结构变异转录本进行酵母转录自激活试验,研究序列结构变异对TaNAC基因转录调控活性的影响。此外,利用miRBase数据库收录的小麦中已报道的miRNAs和TaNAC基因,进行靶基因预测分析,建立小麦TaNAC家族成员与miRNAs的靶向关系。【结果】以条锈菌和白粉菌侵染的普通小麦兼抗种质N9134为材料,克隆得到的TaNAC转录本中的35条序列结构变异转录本由13个TaNAC基因可变剪切形成。通过分析发现,同一TaNAC基因由可变剪切形成的不同结构变异转录本的核酸序列结构存在差异,而且其对应编码产物的功能结构域、高级结构、理化性质和亚细胞定位等方面均会存在差异,同时也会表现出不同的转录调控活性;不同TaNAC基因的可变剪切方式存在差异,而且它们的结构变异转录本及其编码产物在结构特征、理化性质和转录调控活性等方面也均呈现出多样性的特征。通过分析TaNAC基因与其在编码序列区域的靶标tae-miRNAs,发现具有可变剪切转录本的TaNAC基因与tae-miRNA的结合位点均在其非可变剪切区域。【结论】TaNAC基因可以通过可变剪切这种转录后调控方式参与小麦对真菌胁迫的响应;同时发现,调控TaNAC基因的tae-miRNA可以独立于可变剪切这种转录后调控方式行使功能。
吕士凯, 马小龙, 张敏, 邓平川, 陈春环, 张宏, 刘新伦, 吉万全. 小麦TaNAC基因基于可变剪切和microRNA的转录后调控分析[J]. 中国农业科学, 2021, 54(22): 4709-4727.
LÜ ShiKai, MA XiaoLong, ZHANG Min, DENG PingChuan, CHEN ChunHuan, ZHANG Hong, LIU XinLun, JI WanQuan. Post-transcriptional Regulation of TaNAC Genes by Alternative Splicing and MicroRNA in Common Wheat (Triticum aestivum L.)[J]. Scientia Agricultura Sinica, 2021, 54(22): 4709-4727.
表1
TaNAC可变剪切结构变异转录本的克隆引物序列"
| 分组 Groups | TaNAC基因 TaNAC genes | 正向引物序列 Forward primer sequences (5′-3′) | 反向引物序列 Reversed primer sequences (5′-3′) |
|---|---|---|---|
| 1 | TaNAC_NTL5_7B | ATGACCCACCCTTCCTCGTC | TTACTTGCCGTAGATGCACAG |
| 2 | TaNAC008_3A | ATGGAAACTCTACGTGACATGGCT | CTACAAACTGATCCCACCACGGT |
| 3 | TaNAC013_3A | ATGACATGGTGCAATAGCTTC | TCAGGGACCAAAGCCCGTC |
| 4 | TaNAC017_5A | ATGGAGACCCCGCCGCCG | TCATGTAGAGGACTTCCATAAC |
| 5 | TaNAC011_7D | ATGGACGACGGTGCAACG | TCACCCATGATGATCCTGGT |
| 6 | TaNAC017_5D | ATGGAGACCCCGCCGCCG | CTTTGTGCATCATCTTGTCATG |
| 7 | TaNAC024_3D | ATGAACAGGGGTCACATCAGC | CTATCCATGATGATCTTGGTTGTC |
| 8 | TaNAC039_6B | ATGGACACTTTCTCCCATGTTC | CTATTTCCACAGATGAGCCGAG |
| 9 | TaNAC073_3B | ATGGACAAGGGTCACCCAGGGTAC | CTAGTACTGGTAAGGCCATTGCAC |
| 10 | TaNAC092_2A | TCCAGACGGCAAATTCCAG | TCAGTAGCCCCACGGCGC |
| 11 | TaNAC092_2D | ATGGAGCACAGCGAGCAGG | TCAGTAGCCCCACGGCGC |
| 12 | TaNAC024_3A | ATGAACAGGGGTCACATCAGC | CTATGAGATCTCCCGGTTCTTCTC |
| 13 | TaNAC092_2B | ATGGAGCACAGCGAGCAGG | AGCTTGCTTGGTCAGTAGCC |
表2
克隆得到的13个基因可变剪切形成的35条TaNAC结构变异转录本"
| 分组 Groups | TaNAC转录本名称 Names of TaNAC transcripts | 序列长度 Sequence length (bp) | GenBank登录号 Accessions | 对应中国春参考基因组的基因 The corresponding genes in IWGSC RefSeq v1.1 | 是否外显子跳跃 Exon skipping or not | 是否内含子保留 Intron retention or not | 转录本编码方式 Coding mode of transcripts |
|---|---|---|---|---|---|---|---|
| 1 | TaNAC_NTL5_7B.2 | 1932 | MN747301 | TraesCS7B02G196900 | 是 Yes | 否 No | 正常 N |
| TaNAC_NTL5_7B.3 | 879 | MN747304 | TraesCS7B02G196900 | 正常 N | |||
| 2 | TaNAC008_3A.1 | 1446 | MN747252 | TraesCS3A02G176500 | 是 Yes | 否 No | 正常 N |
| TaNAC008_3A.2 | 597 | MN747253 | TraesCS3A02G176500 | 正常 N | |||
| 3 | TaNAC013_3A.1 | 1017 | MN747260 | TraesCS3A02G245900 | 是 Yes | 否 No | 正常 N |
| TaNAC013_3A.2 | 1017 | MN747262 | TraesCS3A02G245900 | 正常 N | |||
| TaNAC013_3A.3 | 974 | MN786410 | TraesCS3A02G245900 | 提前终止 P | |||
| 4 | TaNAC017_5A.2 | 2193 | MN747191 | TraesCS5A02G271500 | 是 Yes | 否 No | 正常 N |
| TaNAC017_5A.3 | 1701 | MN747193 | TraesCS5A02G271500 | 正常 N | |||
| TaNAC017_5A.4 | 1181 | MN786415 | TraesCS5A02G271500 | 分段 S | |||
| TaNAC017_5A.5 | 1237 | MN786416 | TraesCS5A02G271500 | 分段 S | |||
| TaNAC017_5A.6 | 1308 | MN747194 | TraesCS5A02G271500 | 正常 N | |||
| 5 | TaNAC011_7D.1 | 1467 | MN747256 | TraesCS7D02G365200 | 否 No | 是 Yes | 正常 N |
| TaNAC011_7D.2 | 865 | MN786437 | TraesCS7D02G365200 | 分段 S | |||
| 6 | TaNAC017_5D.2 | 2160 | MN747189 | TraesCS5D02G279100 | 否 No | 是 Yes | 正常 N |
| TaNAC017_5D.4 | 2052 | IWGSC | TraesCS5D02G279100 | 正常 N | |||
| 7 | TaNAC024_3D.1 | 1320 | MN747206 | TraesCS3D02G109400 | 否 No | 是 Yes | 正常 N |
| TaNAC024_3D.2 | 1317 | MN747210 | TraesCS3D02G109400 | 正常 N | |||
| TaNAC024_3D.3 | 1457 | MN786432 | TraesCS3D02G109400 | 分段 S | |||
| 8 | TaNAC039_6B.3 | 1056 | MN747281 | TraesCS6B02G286200 | 否 No | 是 Yes | 正常 N |
| TaNAC039_6B.5 | 1171 | MN786440 | TraesCS6B02G286200 | 分段 S | |||
| 9 | TaNAC073_3B.3 | 966 | MN747291 | TraesCS3B02G410500 | 否 No | 是 Yes | 正常 N |
| TaNAC073_3B.4 | 1134 | MN786442 | TraesCS3B02G410500 | 分段 S | |||
| TaNAC073_3B.6 | 979 | MN786412 | TraesCS3B02G410500 | 提前终止 P | |||
| 10 | TaNAC092_2A.3 | 1068 | MN747237 | TraesCS2A02G338300 | 否 No | 是 Yes | 正常 N |
| TaNAC092_2A.5 | 1325 | MN786430 | TraesCS2A02G338300 | 分段 S | |||
| 11 | TaNAC092_2D.6 | 1062 | MN747248 | TraesCS2D02G324700 | 否 No | 是 Yes | 正常 N |
| TaNAC092_2D.7 | 1403 | MN786434 | TraesCS2D02G324700 | 分段 S | |||
| TaNAC092_2D.8 | 1405 | MN786435 | TraesCS2D02G324700 | 分段 S | |||
| 12 | TaNAC024_3A.1 | 1320 | MN747211 | TraesCS3A02G107400 | 是 Yes | 是 Yes | 正常 N |
| TaNAC024_3A.3 | 1317 | MN747208 | TraesCS3A02G107400 | 正常 N | |||
| TaNAC024_3A.6 | 1612 | MN786431 | TraesCS3A02G107400 | 分段 S | |||
| TaNAC024_3A.7 | 1401 | MN786419 | TraesCS3A02G107400 | 分段 S | |||
| 13 | TaNAC092_2B.1 | 1065 | MN747239 | TraesCS2B02G343600 | 是 Yes | 是 Yes | 正常 N |
| TaNAC092_2B.3 | 708 | MN747234 | TraesCS2B02G343600 | 正常 N | |||
| TaNAC092_2B.7 | 1167 | MN786429 | TraesCS2B02G343600 | 分段 S |
表3
克隆得到的13组TaNAC结构变异转录本中正常编码产物的结构特征和理化性质"
| 分组 Groups | 克隆的转录本名称 Name of cloned transcripts | 肽链长度 Peptide length (aa) | NAM结构域 起止位置 NAM start_end (aa) | 跨膜结构域位置 Topology of TMHs | 脂肪系数 Aliphatic index | 亲水性总 平均值 GRAVY | 不稳定系数 Instability index | 理论等电点 pI | 预测的亚细胞定位 Predicted subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TaNAC_NTL5_7B.2 | 643 | 23—149 | o620—642i | 71.12 | -0.424 | 36.32 | 4.63 | 细胞核 Nu |
| TaNAC_NTL5_7B.3 | 292 | 无 None | o269—291i | 70.45 | -0.114 | 40.41 | 4.61 | 胞外/细胞核 Ex / Nu | |
| 2 | TaNAC008_3A.1 | 481 | 10—136 | o448—470i | 72.45 | -0.554 | 35.92 | 5.67 | 细胞核/细胞质 Nu / Cy |
| TaNAC008_3A.2 | 198 | 10—136 | 无 None | 65.56 | -0.809 | 37.89 | 9.08 | 细胞质 Cy | |
| 3 | TaNAC013_3A.1 | 338 | 73—212 | 无 None | 63.08 | -0.656 | 35.37 | 8.83 | 细胞核/线粒体 Nu / Mi |
| TaNAC013_3A.2 | 338 | 73—212 | 无 None | 63.08 | -0.655 | 35.48 | 8.83 | 细胞核/线粒体/细胞质 Nu / Mi / Cy | |
| TaNAC013_3A.3 | 266 | 73—212 | 无 None | 67.18 | -0.676 | 36.58 | 9.15 | 细胞核/线粒体 Nu / Mi | |
| 4 | TaNAC017_5A.2 | 730 | 13—136 | o695—717i | 69.74 | -0.511 | 49.35 | 4.55 | 细胞核 Nu |
| TaNAC017_5A.3 | 566 | 无 None | o531—553i | 72.86 | -0.403 | 51.11 | 4.18 | 细胞核/胞外 Nu / Ex | |
| TaNAC017_5A.6 | 435 | 13—136 | o400—422i | 76.00 | -0.476 | 50.17 | 5.92 | 细胞核 Nu | |
| TaNAC017_5A.4 | 分段 S | — — — — — — — — — — — — | |||||||
| TaNAC017_5A.5 | 分段 S | — — — — — — — — — — — — | |||||||
| 5 | TaNAC011_7D.1 | 488 | 57—226 | 无 None | 53.79 | -0.843 | 55.96 | 7.53 | 细胞核 Nu |
| TaNAC011_7D.2 | 分段 S | — — — — — — — — — — — — | |||||||
| 6 | TaNAC017_5D.2 | 719 | 13—136 | o683—705i | 69.61 | -0.490 | 49.99 | 4.57 | 细胞核 Nu |
| TaNAC017_5D.4* | 683 | 13—136 | o647—669i | 70.26 | -0.528 | 50.36 | 4.62 | 细胞核 Nu | |
| 7 | TaNAC024_3D.1 | 439 | 45—186 | 无 None | 59.13 | -0.813 | 48.33 | 6.88 | 细胞核 Nu |
| TaNAC024_3D.2 | 438 | 44—185 | 无 None | 59.27 | -0.806 | 48.24 | 6.76 | 细胞核 Nu | |
| TaNAC024_3D.3 | 分段 S | — — — — — — — — — — — — | |||||||
| 8 | TaNAC039_6B.3 | 351 | 8—135 | 无 None | 58.97 | -0.755 | 38.99 | 6.57 | 细胞核 Nu |
| TaNAC039_6B.5 | 分段 S | — — — — — — — — — — — — | |||||||
| 9 | TaNAC073_3B.3 | 321 | 46—188 | 无 None | 54.7 | -0.902 | 37.18 | 8.6 | 细胞核 Nu |
| TaNAC073_3B.6 | 278 | 46—188 | 无 None | 59.64 | -0.828 | 33.43 | 8.83 | 细胞核 Nu | |
| TaNAC073_3B.4 | 分段 S | — — — — — — — — — — — — | |||||||
| 10 | TaNAC092_2A.3 | 355 | 14—138 | 无 None | 64.14 | -0.412 | 30.91 | 7.21 | 细胞核/叶绿体 Nu / Ch |
| TaNAC092_2A.5 | 分段 S | — — — — — — — — — — — — | |||||||
| 11 | TaNAC092_2D.6 | 353 | 14—138 | 无 None | 65.33 | -0.397 | 33.56 | 6.86 | 细胞核/叶绿体 Nu / Ch |
| TaNAC092_2D.7 | 分段 S | — — — — — — — — — — — — | |||||||
| TaNAC092_2D.8 | 分段 S | — — — — — — — — — — — — | |||||||
| 12 | TaNAC024_3A.1 | 439 | 45—186 | 无 None | 58.45 | -0.828 | 46.99 | 6.76 | 细胞核 Nu |
| TaNAC024_3A.3 | 438 | 44—185 | 无 None | 58.58 | -0.821 | 46.9 | 6.67 | 细胞核 Nu | |
| TaNAC024_3A.6 | 分段 S | — — — — — — — — — — — — | |||||||
| TaNAC024_3A.7 | 分段 S | — — — — — — — — — — — — | |||||||
| 13 | TaNAC092_2B.1 | 354 | 14—138 | 无 None | 65.14 | -0.386 | 31.18 | 6.67 | 叶绿体/细胞核 Ch / Nu |
| TaNAC092_2B.3 | 235 | 14—136 | 无 None | 57.87 | -0.506 | 31.91 | 5.27 | 胞外/细胞核 Ex / Nu | |
| TaNAC092_2B.7 | 分段 S | — — — — — — — — — — — — | |||||||
表4
部分TaNAC可变剪切结构变异转录本正常编码产物二级结构的比较"
| 分组Groups | 转录本 Transcripts | 肽链长度 Peptide length (aa) | α螺旋中氨基酸的数量 Number of aa in alpha helix | α螺旋中氨基酸的比例 Percentage of aa in alpha helix (%) | β折叠中氨基酸的数量 Number of aa in beta turn | β折叠中氨基酸的比例 Percentage of aa in beta turn (%) | 延伸链中氨基酸的数量 Number of aa in extended strand | 延伸链中氨基酸的比例 Percentage of aa in extended strand (%) | 无规则卷曲中氨基酸的数量 Number of aa in random coil | 无规则卷曲中 氨基酸的比例 Percentage of aa in random coil (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TaNAC_NTL5_7B.2 | 643 | 140 | 21.77 | 25 | 3.89 | 95 | 14.77 | 383 | 59.56 |
| TaNAC_NTL5_7B.3 | 292 | 51 | 17.47 | 13 | 4.45 | 45 | 15.41 | 183 | 62.67 | |
| 2 | TaNAC008_3A.1 | 481 | 137 | 28.48 | 25 | 5.20 | 60 | 12.47 | 259 | 53.85 |
| TaNAC008_3A.2 | 198 | 29 | 14.65 | 12 | 6.06 | 43 | 21.72 | 114 | 57.58 | |
| 3 | TaNAC013_3A.1 | 338 | 82 | 24.26 | 29 | 8.58 | 66 | 19.53 | 161 | 47.63 |
| TaNAC013_3A.2 | 338 | 89 | 26.33 | 19 | 5.62 | 64 | 18.93 | 166 | 49.11 | |
| TaNAC013_3A.3 | 266 | 61 | 22.93 | 17 | 6.39 | 57 | 21.43 | 131 | 49.25 | |
| 4 | TaNAC017_5A.2 | 730 | 213 | 29.18 | 60 | 8.22 | 106 | 14.52 | 351 | 48.08 |
| TaNAC017_5A.3 | 566 | 163 | 28.80 | 35 | 6.18 | 85 | 15.02 | 283 | 50.00 | |
| TaNAC017_5A.6 | 435 | 121 | 27.82 | 37 | 8.51 | 75 | 17.24 | 202 | 46.44 | |
| 6 | TaNAC017_5D.2 | 719 | 217 | 30.18 | 54 | 7.51 | 99 | 13.77 | 349 | 48.54 |
| TaNAC017_5D.4 | 683 | 198 | 28.99 | 50 | 7.32 | 97 | 14.20 | 338 | 49.49 | |
| 9 | TaNAC073_3B.3 | 321 | 76 | 23.68 | 24 | 7.48 | 56 | 17.45 | 165 | 51.40 |
| TaNAC073_3B.6 | 278 | 63 | 22.66 | 32 | 11.51 | 54 | 19.42 | 129 | 46.40 | |
| 13 | TaNAC092_2B.1 | 354 | 67 | 18.93 | 15 | 4.24 | 46 | 12.99 | 226 | 63.84 |
| TaNAC092_2B.3 | 235 | 87 | 37.02 | 21 | 8.94 | 26 | 11.06 | 101 | 42.98 |

图6
部分TaNAC基因的结构变异转录本转录调控活性的比较分析 a:连接不同目的片段的重组pGBKT7载体转化至酵母菌株Y2H Gold的阳性菌株在SD/-Trp/X-α-Gal固体培养基的生长情况;b:TaNAC008_3A的2个结构变异转录本及阴阳性对照重组载体对应的酵母菌株在SD/-Trp、SD/-Trp/-His/-Ade和SD/-Trp/AbA固体培养基的生长情况;a和b中的A—E为样本的行代号,1—4为样本的列代号,行列代号组合后的“A1—E1”代表转化有连接着不同目的片段的重组pGBKT7载体的Y2H Gold阳性酵母菌株,具体见表5;E2:转化了pGBKT7-53+pGADT7-T载体的阳性对照组酵母菌株,E3:转化有pGBKT7空载体的阴性对照组酵母菌株"
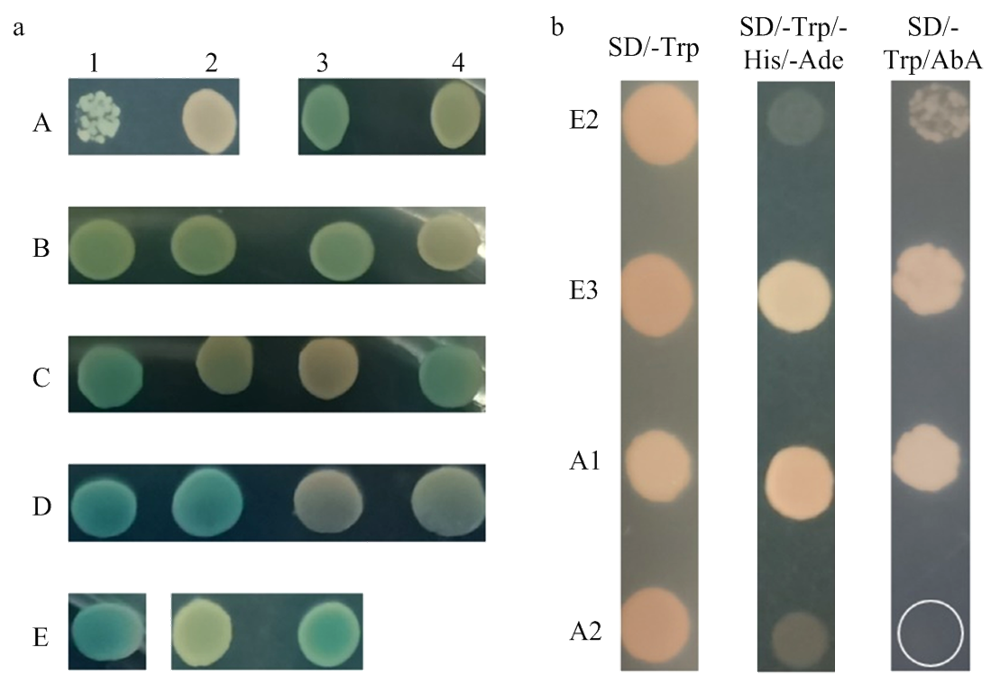
表5
5组TaNAC基因的17条结构变异转录本的转录调控活性分析结果"
Number in | 转录本名称 Transcripts | 插入片段的长度 Length of insert fragment (bp) | 是否具有激活功能 Activation or not | |||
|---|---|---|---|---|---|---|
| SD/-Trp | SD/-Trp/X-α-gal | SD/-Trp/AbA | SD/-Trp/-His/-Ade | |||
| A1 | TaNAC008_3A.1 | 1446 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| A2 | TaNAC008_3A.2 | 597 | 是 Yes | 否 No | 否 No | 否 No |
| A3 | TaNAC_NTL5_7B.2 | 1932 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| A4 | TaNAC_NTL5_7B.3 | 879 | 是 Yes | 否 No | 是 Yes | 是 Yes |
| B1 | TaNAC013_3A.3 | 801/974 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| B2 | TaNAC013_3A.3 | 974 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| B3 | TaNAC013_3A.1 | 1017 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| B4 | TaNAC013_3A.2 | 1017 | 是 Yes | 否 No | 是 Yes | 是 Yes |
| C1 | TaNAC092_2A.3 | 1068 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| C2 | TaNAC092_2A.5 | 1175 | 是 Yes | 否 No | 是 Yes | 是 Yes |
| C3 | TaNAC092_2A.5 | 633(1—633)/1175 | 是 Yes | 否 No | 是 Yes | 是 Yes |
| C4 | TaNAC092_2A.5 | 483(693—1175)/1175 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| D1 | TaNAC092_2B.1 | 1065 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| D2 | TaNAC092_2B.3 | 708 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
| D3 | TaNAC092_2B.7 | 1167 | 是 Yes | 否 No | 是 Yes | 是 Yes |
| D4 | TaNAC092_2B.7 | 555(1—555)/1167 | 是 Yes | 否 No | 是 Yes | 是 Yes |
| E1 | TaNAC092_2B.7 | 483(685—1167)/1167 | 是 Yes | 是 Yes | 是 Yes | 是 Yes |
表6
小麦参考基因组(IWGSC RefSeq v1.1)注释的TaNAC转录本与小麦tae-miRNA的靶向关系预测结果"
| miRNA名称 miRNA ID | 期望值 Expectation | 抑制方式 Inhibition | TaNAC转录本名称 TaNAC transcripts ID | 靶基因 长度 Length (bp) | 是否可 变剪切 AS or not | NAM结构域编码起止位置 NAM coding start_end (bp) | miRNA结合起止位置 Target miRNA start_end (bp) | 靶基因与miRNA匹配的序列 Target aligned fragment (5′-3′) | 匹配方式 Alignment |
|---|---|---|---|---|---|---|---|---|---|
| tae-miR164 | 1 | 裂解 Cleavage | TraesCS5A02G049100.1 | 918 | 是 Yes | 40—423 | 653—673 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: |
| TraesCS5A02G049100.2 | 921 | 是 Yes | 40—423 | 656—676 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS5B02G054200.1 | 921 | 是 Yes | 40—423 | 656—676 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS5B02G054200.2 | 924 | 是 Yes | 40—423 | 659—679 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS5D02G059700.1 | 909 | 是 Yes | 40—423 | 644—664 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS5D02G059700.2 | 912 | 是 Yes | 40—423 | 647—667 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS7A02G334800.1 | 1092 | 否 No | 64—444 | 770—790 | AGCUCGUGCCCUGCUUCUCCA | :: ::::::::::::::::: | |||
| TraesCS7A02G464100.1 | 924 | 否 No | 34—435 | 653—673 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS7A02G464800.1 | 927 | 否 No | 34—438 | 656—676 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS7B02G246300.1 | 1089 | 否 No | 55—444 | 767—787 | AGCUCGUGCCCUGCUUCUCCA | :: ::::::::::::::::: | |||
| TraesCS7B02G364600.1 | 924 | 否 No | 34—435 | 653—673 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS7B02G365300.1 | 918 | 否 No | 34—429 | 647—667 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS7D02G342300.1 | 1089 | 否 No | 64—444 | 767—787 | AGCUCGUGCCCUGCUUCUCCA | :: ::::::::::::::::: | |||
| TraesCS7D02G451700.1 | 939 | 否 No | 34—438 | 656—676 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| TraesCS7D02G452500.1 | 927 | 否 No | 34—435 | 656—676 | AGCAAGUGCCCUGCUUCUCCA | ::: :::::::::::::::: | |||
| 1.5 | 裂解 Cleavage | TraesCS2A02G338300.1 | 1068 | 否 No | 40—414 | 680—700 | CGCACGUGACCUGCUUCUCCA | ::::::: :::::::::::: | |
| TraesCS2B02G343600.1 | 1065 | 否 No | 40—414 | 680—700 | CGCACGUGACCUGCUUCUCCA | ::::::: :::::::::::: | |||
| TraesCS2D02G324700.1 | 1062 | 否 No | 40—414 | 677—697 | CGCACGUGACCUGCUUCUCCA | ::::::: :::::::::::: | |||
| 2 | 裂解 Cleavage | TraesCS7A02G305200.1 | 870 | 否 No | 34—414 | 665—685 | AGCAAGUGUCCUGCUUCUCCG | ::: :::.:::::::::::. | |
| TraesCS7B02G205600.1 | 879 | 否 No | 34—414 | 674—694 | AGCAAGUGUCCUGCUUCUCCG | ::: :::.:::::::::::. | |||
| TraesCS7D02G302000.1 | 876 | 否 No | 34—420 | 671—691 | AGCAAGUGUCCUGCUUCUCCG | ::: :::.:::::::::::. | |||
| miRNA名称 miRNA ID | 期望值 Expectation | 抑制方式 Inhibition | TaNAC转录本名称 TaNAC transcripts ID | 靶基因 长度 Length (bp) | 是否可 变剪切 AS or not | NAM结构域编码起止位置 NAM coding start_end (bp) | miRNA结合起止位置 Target miRNA start_end (bp) | 靶基因与miRNA匹配的序列 Target aligned fragment (5′-3′) | 匹配方式 Alignment |
| tae-miR1128 | 3.5 | 裂解 Cleavage | TraesCS3A02G339600.1 | 945 | 是 Yes | 97—480 | 246—266 | GUUCGGGAGCAGGGAGUGGUA | :::::. : :::::::.::: |
| TraesCS3A02G339600.2 | 882 | 是 Yes | 97—651 | 371—391 | GUUCGGGAGCAGGGAGUGGUA | :::::. : :::::::.::: | |||
| TraesCS3B02G371200.1 | 939 | 否 No | 85—468 | 234—254 | GUUCGGGAGCAGGGAGUGGUA | :::::. : :::::::.::: | |||
| TraesCS3D02G333100.1 | 951 | 否 No | 97—480 | 246—266 | GUUCGGGAGCAGGGAGUGGUA | :::::. : :::::::.::: | |||
| TraesCS4B02G328800.1 | 1044 | 否 No | 34—414 | 180—200 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS4B02G328900.1 | 1059 | 否 No | 49—429 | 195—215 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS4B02G329100.1 | 1059 | 否 No | 52—432 | 198—218 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS4D02G325800.1 | 1065 | 否 No | 52—432 | 198—218 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS5A02G500400.1 | 1017 | 否 No | 22—402 | 195—215 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS5A02G500500.1 | 1110 | 否 No | 52—432 | 168—188 | AAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS5A02G500600.1 | 1071 | 否 No | 52—432 | 198—218 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| TraesCS5A02G500700.1 | 2085 | 否 No | 19—399 | 198—218 | GAUCGGGGAGAGGGAGUGGUA | ::::. ::::::::.::: | |||
| tae-miR408 | 3 | 翻译 Translation | TraesCS6A02G378500.1 | 936 | 否 No | 31—441 | 812—832 | CCCAGGGCGGCGGCAGUGCAG | ::::::.: :::::::::: |
| 3.5 | 裂解 Cleavage | TraesCS4B02G320300.1 | 1104 | 否 No | 25—408 | 622—642 | UUCAAGGAGGAGGCGGAGCAG | .:: :::.:::::.: :::: | |
| TraesCS4D02G316800.1 | 1104 | 否 No | 25—408 | 622—642 | UUCAAGGAGGAGGCGGAGCAG | .:: :::.:::::.: :::: | |||
| TraesCS5A02G491700.1 | 1047 | 否 No | 49—429 | 622—642 | UUCAAGGAGGAGGCGGAGCAG | .:: :::.:::::.: :::: | |||
| tae-miR9780 | 3 | 翻译 Translation | TraesCSU02G120000.1 | 1239 | 否 No | 22—450 | 136—156 | GACGCGUACGCCGCCGACCCG | : ::::: :. :::::::::: |
| 3.5 | 裂解 Cleavage | TraesCS6A02G065600.1 | 1227 | 否 No | 22—444 | 136—156 | GACGCGUACGGCGCAGAUCCG | : ::::: :.:::: ::.::: | |
| 翻译 Translation | TraesCSU02G119900.1 | 1284 | 否 No | 22—450 | 136—156 | GACGCGUACGCCGCCGAUCCG | : ::::: :. ::::::.::: | ||
| tae-miR9676-5p | 2.5 | 裂解 Cleavage | TraesCS5B02G480900.1 | 900 | 是 Yes | 58—351 | 755—776 | ACCUCAGCUACGAUGACAUCCA | : ::.::::::::::::: |
| TraesCS5B02G480900.2 | 984 | 是 Yes | 58—435 | 839—860 | ACCUCAGCUACGAUGACAUCCA | : ::.::::::::::::: | |||
| tae-miR171a | 3.5 | 裂解 Cleavage | TraesCS4D02G175700.1 | 1074 | 否 No | 28—408 | 747—767 | GCUGAUGGCACGGCACGAUCA | : :. ::::::::: :.:::: |
| tae-miR444a | 3.5 | 翻译 Translation | TraesCS7A02G349500.1 | 1107 | 否 No | 31—420 | 1011—1031 | GGAGCAACUUGGAGGCAGCAG | : :::::.: :::::::::. |
| tae-miR444b | 3.5 | 翻译 Translation | TraesCS7A02G349500.1 | 1107 | 否 No | 31—420 | 1011—1031 | GGAGCAACUUGGAGGCAGCAG | : :::::.: :::::::::. |
| tae-miR9677a | 3.5 | 裂解 Cleavage | TraesCS1A02G276000.1 | 774 | 否 No | 13—405 | 296—317 | UGGUCUACUCCGCCGGCGGCCA | .::::::.::..:::::: |
表7
小麦、玉米、水稻、二穗短柄草和拟南芥中已报道miRNAs的汇总分析"
| 植物 Plants | miRNA前体数量 Number of miRNA precursors | 成熟miRNA数量 Number of mature miRNAs | miRNA家族数量 Number of miRNA families | 基因组大小 Genome sizes (G) |
|---|---|---|---|---|
| 普通小麦 Triticum aestivum | 122 | 125 | 99 | 14.5 |
| 玉米 Zea mays | 174 | 325 | 31 | 2.4 |
| 水稻 Oryza sativa | 604 | 738 | 342 | 0.5 |
| 二穗短柄草 Brachypodium distachyon | 317 | 525 | 183 | 0.385 |
| 拟南芥 Arabidopsis thaliana | 326 | 428 | 215 | 0.13 |
| [1] |
FUJITA M, FUJITA Y, MARUYAMA K, SEKI M, HIRATSU K, OHME-TAKAGI M, TRAN L S, YAMAGUCHI-SHINOZAKI K, SHINOZAKI K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. The Plant Journal, 2004, 39(6): 863-876.
doi: 10.1111/tpj.2004.39.issue-6 |
| [2] |
TRAN L S, NAKASHIMA K, SAKUMA Y, SIMPSON S D, FUJITA Y, MARUYAMA K, FUJITA M, SEKI M, SHINOZAKI K, YAMAGUCHI-SHINOZAKI K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell, 2004, 16(9): 2481-2498.
doi: 10.1105/tpc.104.022699 |
| [3] |
GUO Y, GAN S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal, 2006, 46(4): 601-612.
doi: 10.1111/tpj.2006.46.issue-4 |
| [4] | HU H, DAI M, YAO J, XIAO B, LI X, ZHANG Q, XIONG L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(35): 12987-12992. |
| [5] |
XU B, OHTANI M, YAMAGUCHI M, TOYOOKA K, WAKAZAKI M, SATO M, KUBO M, NAKANO Y, SANO R, HIWATASHI Y, MURATA T, KURATA T, YONEDA A, KATO K, HASEBE M, DEMURA T. Contribution of NAC transcription factors to plant adaptation to land. Science, 2014, 343(6178): 1505-1508.
doi: 10.1126/science.1248417 |
| [6] |
LV S, GUO H, ZHANG M, WANG Q, ZHANG H, JI W. Large-scale cloning and comparative analysis of TaNAC genes in response to stripe rust and powdery mildew in wheat (Triticum aestivum L.). Genes (Basel), 2020, 11(9): 1073.
doi: 10.3390/genes11091073 |
| [7] |
FENG H, DUAN X, ZHANG Q, LI X, WANG B, HUANG L, WANG X, KANG Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Molecular Plant Pathology, 2014, 15(3): 284-296.
doi: 10.1111/mpp.2014.15.issue-3 |
| [8] |
RIECHMANN J L, HEARD J, MARTIN G, REUBER L, JIANG C, KEDDIE J, ADAM L, PINEDA O, RATCLIFFE O J, SAMAHA R R, CREELMAN R, PILGRIM M, BROUN P, ZHANG J Z, GHANDEHARI D, SHERMAN B K, YU G. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science, 2000, 290(5499): 2105-2110.
doi: 10.1126/science.290.5499.2105 |
| [9] |
SOUER E, VANHOUWELINGEN A, KLOOS D, MOL J, KOES R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell, 1996, 85(2): 159-170.
doi: 10.1016/S0092-8674(00)81093-4 |
| [10] |
AIDA M, ISHIDA T, FUKAKI H, FUJISAWA H, TASAKA M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. The Plant Cell, 1997, 9(6): 841-857.
doi: 10.1105/tpc.9.6.841 |
| [11] |
OOKA H, SATOH K, DOI K, NAGATA T, OTOMO Y, MURAKAMI K, MATSUBARA K, OSATO N, KAWAI J, CARNINCI P, HAYASHIZAKI Y, SUZUKI K, KOJIMA K, TAKAHARA Y, YAMAMOTO K, KIKUCHI S. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research, 2003, 10(6): 239-247.
doi: 10.1093/dnares/10.6.239 |
| [12] |
OLSEN A N, ERNST H A, LEGGIO L L, SKRIVER K. NAC transcription factors: Structurally distinct, functionally diverse. Trends in Plant Science, 2005, 10(2): 79-87.
doi: 10.1016/j.tplants.2004.12.010 |
| [13] |
MUROZUKA E, MASSANGE-SANCHEZ J A, NIELSEN K, GREGERSEN P L, BRAUMANN I. Genome wide characterization of barley NAC transcription factors enables the identification of grain-specific transcription factors exclusive for the Poaceae family of monocotyledonous plants. PLoS ONE, 2018, 13(12): e0209769.
doi: 10.1371/journal.pone.0209769 |
| [14] | TIAN F, YANG D C, MENG Y Q, JIN J, GAO G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Research, 2020, 48(D1): D1104-D1113. |
| [15] |
GUERIN C, ROCHE J, ALLARD V, RAVEL C, MOUZEYAR S, BOUZIDI M F. Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS ONE, 2019, 14(3): e0213390.
doi: 10.1371/journal.pone.0213390 |
| [16] |
ZHAO D, DERKX A P, LIU D C, BUCHNER P, HAWKESFORD M J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biology (Stuttgart), 2015, 17(4): 904-913.
doi: 10.1111/plb.2015.17.issue-4 |
| [17] |
HARRINGTON S A, OVEREND L E, COBO N, BORRILL P, UAUY C. Conserved residues in the wheat (Triticum aestivum) NAM-A1 NAC domain are required for protein binding and when mutated lead to delayed peduncle and flag leaf senescence. BMC Plant Biology, 2019, 19(1): 407.
doi: 10.1186/s12870-019-2022-5 |
| [18] |
XUE G P, BOWER N I, MCINTYRE C L, RIDING G A, KAZAN K, SHORTER R. TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Functional Plant Biology, 2006, 33(1): 43-57.
doi: 10.1071/FP05161 |
| [19] | ZHANG L, ZHANG L, XIA C, ZHAO G, JIA J, KONG X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front Plant Science, 2015, 6: 1174. |
| [20] |
PEROCHON A, KAHLA A, VRANIC M, JIA J, MALLA K B, CRAZE M, WALLINGTON E, DOOHAN F M. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnology Journal, 2019, 17(10): 1892-1904.
doi: 10.1111/pbi.v17.10 |
| [21] |
WANG B, WEI J, SONG N, WANG N, ZHAO J, KANG Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. Journal of Integrative Plant Biology, 2018, 60(5): 432-443.
doi: 10.1111/jipb.v60.5 |
| [22] |
PURANIK S, SAHU P P, SRIVASTAVA P S, PRASAD M. NAC proteins: Regulation and role in stress tolerance. Trends in Plant Science, 2012, 17(6): 369-381.
doi: 10.1016/j.tplants.2012.02.004 |
| [23] |
AMERES S L, ZAMORE P D. Diversifying microRNA sequence and function. Nature Reviews. Molecular Cell Biology, 2013, 14(8): 475-488.
doi: 10.1038/nrm3611 |
| [24] |
ZHANG H, MAO R, WANG Y, ZHANG L, WANG C, LV S, LIU X, WANG Y, JI W. Transcriptome-wide alternative splicing modulation during plant-pathogen interactions in wheat. Plant Science, 2019, 288: 110160.
doi: 10.1016/j.plantsci.2019.05.023 |
| [25] |
ZHANG X M, ZHANG Q, PEI C L, LI X, HUANG X L, CHANG C Y, WANG X J, HUANG L L, KANG Z S. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage. Physiological and Molecular Plant Pathology, 2018, 102: 144-153.
doi: 10.1016/j.pmpp.2018.02.002 |
| [26] |
WANG H-L, ZHANG Y, WANG T, YANG Q, YANG Y, LI Z, LI B, WEN X, LI W, YIN W, XIA X, GUO H, LI Z. An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. The Plant Cell, 2021, 33: 1594-1614.
doi: 10.1093/plcell/koab046 |
| [27] |
REDDY A S, MARQUEZ Y, KALYNA M, BARTA A. Complexity of the alternative splicing landscape in plants. The Plant Cell, 2013, 25(10): 3657-3683.
doi: 10.1105/tpc.113.117523 |
| [28] |
XUE F, JI W, WANG C, ZHANG H, YANG B. High-density mapping and marker development for the powdery mildew resistance gene PmAS846 derived from wild emmer wheat (Triticum turgidum var. dicoccoides). Theoretical and Applied Genetics, 2012, 124(8): 1549-1560.
doi: 10.1007/s00122-012-1809-7 |
| [29] |
ZHANG H, YANG Y, WANG C, LIU M, LI H, FU Y, WANG Y, NIE Y, LIU X, JI W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics, 2014, 15(1): 898.
doi: 10.1186/1471-2164-15-898 |
| [30] |
ZHANG H, MAO R, WANG Y, ZHANG L, WANG C, LV S, LIU X, WANG Y, JI W. Transcriptome-wide alternative splicing modulation during plant-pathogen interactions in wheat. Plant Science, 2019, 288: 110160.
doi: 10.1016/j.plantsci.2019.05.023 |
| [31] |
SANCHEZ-MARTIN J, WIDRIG V, HERREN G, WICKER T, ZBINDEN H, GRONNIER J, SPORRI L, PRAZ C R, HEUBERGER M, KOLODZIEJ M C, ISAKSSON J, STEUERNAGEL B, KARAFIATOVA M, DOLEZEL J, ZIPFEL C, KELLER B. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nature Plants, 2021, 7(3): 327-341.
doi: 10.1038/s41477-021-00869-2 |
| [32] |
GAO P, QUILICHINI T D, ZHAI C, QIN L, NILSEN K T, LI Q, SHARPE A G, KOCHIAN L V, ZOU J, REDDY A S N, WEI Y, POZNIAK C, PATTERSON N, GILLMOR C S, DATLA R, XIANG D. Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnology Journal, 2021, 19: 1624-1643.
doi: 10.1111/pbi.v19.8 |
| [33] |
KOZOMARA A, BIRGAOANU M, GRIFFITHS-JONES S. miRBase: From microRNA sequences to function. Nucleic Acids Research, 2019, 47(D1): D155-D162.
doi: 10.1093/nar/gky1141 |
| [34] |
FANG Y, XIE K, XIONG L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. Journal of Experimental Botany, 2014, 65(8): 2119-2135.
doi: 10.1093/jxb/eru072 |
| [35] |
KIM J H, WOO H R, KIM J, LIM P O, LEE I C, CHOI S H, HWANG D, NAM H G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science, 2009, 323(5917): 1053-1057.
doi: 10.1126/science.1166386 |
| [1] | 陈吉浩, 周界光, 曲翔汝, 王素容, 唐华苹, 蒋云, 唐力为, $\boxed{\hbox{兰秀锦}}$, 魏育明, 周景忠, 马建. 四倍体小麦胚大小性状QTL定位与分析[J]. 中国农业科学, 2023, 56(2): 203-216. |
| [2] | 严艳鸽, 张水勤, 李燕婷, 赵秉强, 袁亮. 葡聚糖改性尿素对冬小麦产量和肥料氮去向的影响[J]. 中国农业科学, 2023, 56(2): 287-299. |
| [3] | 徐久凯, 袁亮, 温延臣, 张水勤, 李燕婷, 李海燕, 赵秉强. 畜禽有机肥氮在冬小麦季对化肥氮的相对替代当量[J]. 中国农业科学, 2023, 56(2): 300-313. |
| [4] | 古丽旦,刘洋,李方向,成卫宁. 小麦吸浆虫小热激蛋白基因Hsp21.9的克隆及在滞育过程与温度胁迫下的表达特性[J]. 中国农业科学, 2023, 56(1): 79-89. |
| [5] | 王浩琳,马悦,李永华,李超,赵明琴,苑爱静,邱炜红,何刚,石美,王朝辉. 基于小麦产量与籽粒锰含量的磷肥优化管理[J]. 中国农业科学, 2022, 55(9): 1800-1810. |
| [6] | 唐华苹,陈黄鑫,李聪,苟璐璐,谭翠,牟杨,唐力为,兰秀锦,魏育明,马建. 基于55K SNP芯片的普通小麦穗长非条件和条件QTL分析[J]. 中国农业科学, 2022, 55(8): 1492-1502. |
| [7] | 马小艳,杨瑜,黄冬琳,王朝辉,高亚军,李永刚,吕辉. 小麦化肥减施与不同轮作方式的周年养分平衡及经济效益分析[J]. 中国农业科学, 2022, 55(8): 1589-1603. |
| [8] | 刘硕,张慧,高志源,许吉利,田汇. 437个小麦品种钾收获指数的变异特征[J]. 中国农业科学, 2022, 55(7): 1284-1300. |
| [9] | 王洋洋,刘万代,贺利,任德超,段剑钊,胡新,郭天财,王永华,冯伟. 基于多元统计分析的小麦低温冻害评价及水分效应差异研究[J]. 中国农业科学, 2022, 55(7): 1301-1318. |
| [10] | 职蕾,者理,孙楠楠,杨阳,Dauren Serikbay,贾汉忠,胡银岗,陈亮. 小麦苗期铅耐受性的全基因组关联分析[J]. 中国农业科学, 2022, 55(6): 1064-1081. |
| [11] | 秦羽青,程宏波,柴雨葳,马建涛,李瑞,李亚伟,常磊,柴守玺. 中国北方地区小麦覆盖栽培增产效应的荟萃(Meta)分析[J]. 中国农业科学, 2022, 55(6): 1095-1109. |
| [12] | 蔡苇荻,张羽,刘海燕,郑恒彪,程涛,田永超,朱艳,曹卫星,姚霞. 基于成像高光谱的小麦冠层白粉病早期监测方法[J]. 中国农业科学, 2022, 55(6): 1110-1126. |
| [13] | 宗成, 吴金鑫, 朱九刚, 董志浩, 李君风, 邵涛, 刘秦华. 添加剂对农副产物和小麦秸秆混合青贮发酵品质的影响[J]. 中国农业科学, 2022, 55(5): 1037-1046. |
| [14] | 马鸿翔, 王永刚, 高玉姣, 何漪, 姜朋, 吴磊, 张旭. 小麦抗赤霉病育种回顾与展望[J]. 中国农业科学, 2022, 55(5): 837-855. |
| [15] | 冯子恒,宋莉,张少华,井宇航,段剑钊,贺利,尹飞,冯伟. 基于无人机多光谱和热红外影像信息融合的小麦白粉病监测[J]. 中国农业科学, 2022, 55(5): 890-906. |
|
||