













中国农业科学 ›› 2021, Vol. 54 ›› Issue (20): 4466-4477.doi: 10.3864/j.issn.0578-1752.2021.20.019
收稿日期:2020-09-09
接受日期:2021-05-13
出版日期:2021-10-16
发布日期:2021-10-25
通讯作者:
张红平
作者简介:陈媛,E-mail: 基金资助:
CHEN Yuan( ),CAI He,LI Li,WANG LinJie,ZHONG Tao,ZHANG HongPing(
),CAI He,LI Li,WANG LinJie,ZHONG Tao,ZHANG HongPing( )
)
Received:2020-09-09
Accepted:2021-05-13
Online:2021-10-16
Published:2021-10-25
Contact:
HongPing ZHANG
摘要:
【目的】快速骨骼肌肌钙蛋白T(fast skeletal troponin T3,TNNT3)作为肌钙蛋白(troponin, Tn)家族成员,调节横纹肌收缩、参与骨骼肌的生长发育并影响家畜肉质性状。通过获得山羊TNNT3基因的可变剪切体,分析山羊TNNT3基因可变剪切的表达模式及其在肌细胞分化中的作用,深入解析TNNT3基因在山羊骨骼肌生长发育过程中的作用机制。【方法】基于NCBI已公布山羊TNNT3基因(NM_001314210.1)和牛TNNT3基因(XM_010821200)mRNA序列,使用软件Primer Premier 6.0设计引物,以简州大耳羊胚胎期和出生后7个阶段骨骼肌为试验材料,克隆测序获得山羊TNNT3基因的CDS区可变剪切体,利用软件ORF Finder、EditSeq、DNAMAN、ClustalW和MEGA_X_10.1.8等对序列进行生物信息学分析;进一步设计实时荧光定量(real-time PCR,RT-qPCR)及半定量引物,研究TNNT3基因剪切体在7个不同组织(背最长肌(longissimus dorsi muscle,LD)、半膜肌(semimembranosus muscle,SM)、心、肝、脾、肺、肾)和7个发育阶段(胚胎期E75、E90、E105和出生后B3、B45、B150、B300)肌肉组织(背最长肌和半膜肌)中表达模式;此外,对转录本TNNT3_3进行体外编码能力检测确定其具有编码蛋白的能力,并在山羊骨骼肌卫星细胞(skeletal muscle satellite cells,MuSCs)中过表达,观察细胞形态变化以及检测标志基因的表达变化,研究其对山羊MuSCs分化的作用。【结果】①TNNT3(NM_001314210.1)CDS区全序列主要含有18个外显子,其中外显子16/17相互排斥,转录后单一表达。克隆发现山羊TNNT3基因 5个新转录本(TNNT3_1—5),其外显子数分别是15、15、20、16、14。②生物信息学分析结果显示山羊TNNT3基因核苷酸序列和氨基酸序列与绵羊、牛、猪等哺乳动物具有很高的一致性,而与鱼类和爬行类动物的一致性较低,说明TNNT3基因序列在哺乳动物高度保守。③TNNT3 mRNA在背最长肌、半膜肌、心、肝、脾、肺、肾7个组织中都有表达,其中在骨骼肌中高度富集(P < 0.01),心脏及肺次之,其余组织中较低;TNNT3 mRNA在背最长肌和半膜肌中的表达始终处于一个动态变化中,胚胎期TNNT3在半膜肌的表达量高于背最长肌(P<0.05);出生后则背最长肌中高于半膜肌(P<0.05)。④山羊TNNT3基因转录本TNNT3_3重复出现保守的外显子9—11(138bp),体外翻译实验显示其可编码蛋白且蛋白大小与预期基本相符(37 kD);相较于对照组,在山羊MuSCs中过表达该转录本使肌分化标志基因Myomaker、MyoG和MyH4 mRNA极显著升高(P < 0.01)。【结论】获得了山羊TNNT3基因具有完整CDS区5个新可变剪切体,TNNT3主要在肌肉组织(背最长肌和半膜肌)中高表达,在哺乳动物中高度保守且促进成肌分化。初步表明TNNT3基因在动物肌肉生长发育中具有重要的生物学功能。
陈媛,蔡禾,李利,王林杰,仲涛,张红平. 山羊TNNT3基因可变剪切及其对骨骼肌细胞分化的作用[J]. 中国农业科学, 2021, 54(20): 4466-4477.
CHEN Yuan,CAI He,LI Li,WANG LinJie,ZHONG Tao,ZHANG HongPing. Alternative Splicing of TNNT3 and Its Effect on the Differentiation of MuSCs in Goat[J]. Scientia Agricultura Sinica, 2021, 54(20): 4466-4477.
表1
TNNT3基因引物序列信息"
| 引物名称 Primer name | 引物序列(5′— 3′) Primer sequence (5′— 3′) | 产物长度 Product length (bp) | 退火温度 Annealing temperature (℃) | 用途 Purpose | |
|---|---|---|---|---|---|
| P1 | TNNT3 | F: ACCATGTCGGACGAGGAAGT R: CACTCTACTTCCAGCACCC | 835 | 58.0 | 克隆CDS区全长 Cloning the full length of CDS |
| P2 | q-TNNT3 | F:AGGAGGGCTGAGGACGAT R:CGGTCTCCAGCTTGTAC | 253 | 52.7 | 定量PCR Quantitative PCR |
| P3 | TNNT3_1-5 | F: ACCATGTCGGACGAGGAAGT R:CTCCTTGAGGGCGACCAGC | 253 | 60.0 | 半定量PCR Semi-quantitative PCR |
| P4 | TNT- TNNT3_3 | F:CCCAAGCTTATGTCGGACGAGGAAGTCGA R:CCGCTCGAGCTTCCAGCGCCCGCCAACTT | 957 | 58.9 | TNT体外转录载体构建 Construction of TNT transcriptional vector in vitro |
| P5 | pTNNT3_3 | F:CCGCTCGAGATGTCGGACGAGGAAGTCGA R:CCCAAGCTTCTTCCAGCGCCCGCCAACTT | 957 | 59.2 | 过表达载体构建 Construction of overexpression vectors |
| P6 | β-actin | F:CCTGCGGCATTCACGAAACTAC R:ACAGCACCGTGTTGGCGTAGAG | 87 | 59.7 | 定量PCR Quantitative PCR |

图2
山羊TNNT3基因PCR扩增、克隆及不同可变剪切体序列与山羊全基因组数据库比对 A:TNNT3 CDS区全长PCR产物(P1:835bp);B:TNNT3可变剪切体单克隆菌液PCR产物;C:TNNT3可变剪切体差异片段(exon4-8:193-253 bp)qPCR产物;M1:DNA marker DL 2000 bp;M2: DNA marker DL 5000 bp;D:TNNT3不同可变剪切体序列与山羊全基因组数据库比对结果,红色方框是发生可变剪切的外显子位点;E:山羊TNNT3不同可变剪切体的氨基酸序列对比;绿色方框:骨骼肌中最先被降解的产物[18];红色方框:TNNT3_3的外显子Exon 9—11重复,增加了46个氨基酸残基;蓝色方框:互斥表达的外显子16/17编码的氨基酸[25];F:TNNT3可变剪切体结构域预测"
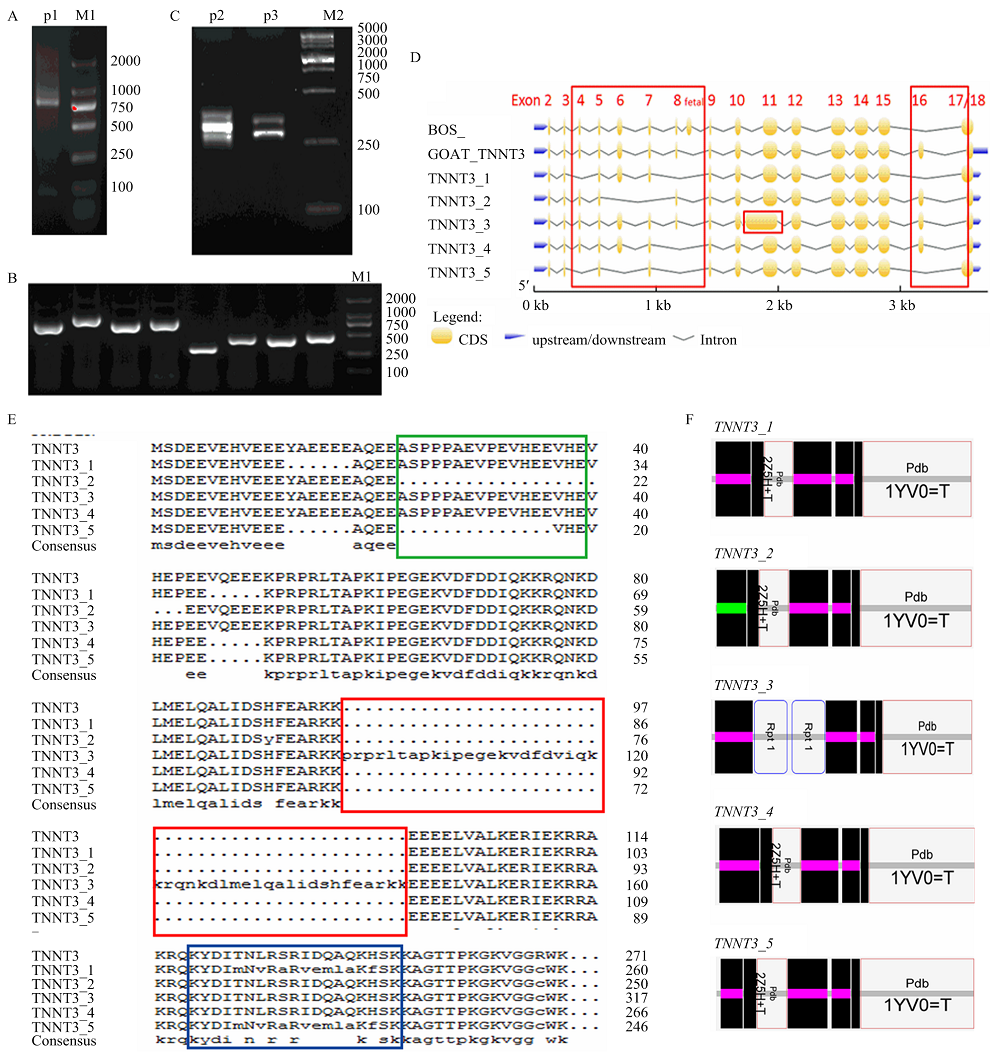
表2
山羊TNNT3不同可变剪切体预测的氨基酸序列信息"
| 序列名称 Sequence name | CDS区长度 Length of CDS (nt) | 外显子数 Exon count | 氨基酸长度 Amino acid length (AA) | 蛋白质分子量 Protein molecular weight (kD) | 等电点 Isoelectric point |
|---|---|---|---|---|---|
| TNNT3_1 | 783 | 15 | 260 | 30.64 | 8.61 |
| TNNT3_2 | 753 | 15 | 250 | 29.73 | 8.30 |
| TNNT3_3 | 954 | 20 | 317 | 37.40 | 6.77 |
| TNNT3_4 | 801 | 16 | 266 | 31.39 | 6.72 |
| TNNT3_5 | 741 | 14 | 246 | 29.17 | 9.15 |
| [1] |
BARTON P J R, MULLEN A J, CULLEN M E, DHOOT G K, SIMON-CHAZOTTES D, GUÉNET J L. Genes encoding troponin I and troponin T are organized as three paralogous pairs in the mouse genome. Mammalian Genome, 2000, 11(10):926-929. doi: 10.1007/s003350010171.
doi: 10.1007/s003350010171 |
| [2] |
GONG H Y, HATCH V, ALI L, LEHMAN W, CRAIG R, TOBACMAN L S. Mini-thin filaments regulated by troponin-tropomyosin. PNAS, 2005, 102(3):656-661. doi: 10.1073/pnas.0407225102.
doi: 10.1073/pnas.0407225102 |
| [3] |
CHEN H, ZHANG J, YU B, LI L, SHANG Y. Molecular cloning, structural analysis, and tissue expression of the TNNT3 gene in Guizhou black goat. Gene, 2015. 573(1):123-128.
doi: 10.1016/j.gene.2015.07.038 |
| [4] |
EBASHI S. Third component participating in the superprecipitation of ‘natural actomyosin’. Nature, 1963, 200:1010. doi: 10.1038/ 2001010a0.
doi: 10.1038/ 2001010a0 |
| [5] |
OTSUKI I, MASAKI T, NONOMURA Y, EBASHI S. Periodic distribution of troponin along the thin filament. Journal of Biochemistry, 1967, 61(6):817-819. doi: 10.1093/oxfordjournals. jbchem.a128619.
doi: 10.1093/oxfordjournals. jbchem.a128619 |
| [6] |
STEFANCSIK R, RANDALL J D, MAO C, SARKAR S. Structure and sequence of the human fast skeletal troponin T (TNNT3) gene: insight into the evolution of the gene and the origin of the developmentally regulated isoforms. Comparative and Functional Genomics, 2003, 4(6):609-625. doi: 10.1002/cfg.343.
doi: 10.1002/cfg.343 |
| [7] |
FLICKER P F, PHILLIPS G N, Jr COHEN C. Troponin and its interactions with tropomyosin. An electron microscope study. Journal of Molecular Biology, 1982. 162(2):495-501.
doi: 10.1016/0022-2836(82)90540-X |
| [8] |
CHAUDHURI T, MUKHERJEA M, SACHDEV S, RANDALL J D, SARKAR S. Role of the fetal and alpha/beta exons in the function of fast skeletal troponin T isoforms: correlation with altered Ca2+ regulation associated with development. Journal of Molecular Biology, 2005, 352(1):58-71. doi: 10.1016/j.jmb.2005.06.066.
doi: 10.1016/j.jmb.2005.06.066 |
| [9] |
BLACK A J, RAVI S, JEFFERSON L S, KIMBALL S R, SCHILDER R J. Dietary fat quantity and type induce transcriptome-wide effects on alternative splicing of pre-mRNA in rat skeletal muscle. The Journal of Nutrition, 2017, 147(9):1648-1657. doi: 10.3945/jn.117. 254482.
doi: 10.3945/jn.117. 254482 |
| [10] | JU Y, LI J, XIE C, RITCHLIN C T, XING L, HILTON M J, SCHWARZ E M. Troponin T3 expression in skeletal and smooth muscle is required for growth and postnatal survival: characterization of Tnnt3(tm2a(KOMP)Wtsi) mice. Genesis, 2013. 51(9):667-675. |
| [11] |
BLACK A J, SCHILDER R J, KIMBALL S R. Palmitate-and C6 ceramide-induced Tnnt3 pre-mRNA alternative splicing occurs in a PP2A dependent manner. Nutrition & Metabolism, 2018, 15:87. doi: 10.1186/s12986-018-0326-3.
doi: 10.1186/s12986-018-0326-3 |
| [12] |
LEE Y, RIO D C. Mechanisms and regulation of alternative pre-mRNA splicing. Annual Review of Biochemistry, 2015, 84:291-323. doi: 10.1146/annurev-biochem-060614-034316.
doi: 10.1146/annurev-biochem-060614-034316 |
| [13] |
NARO C, SETTE C. Phosphorylation-mediated regulation of alternative splicing in cancer. International Journal of Cell Biology, 2013, 2013:151839. doi: 10.1155/2013/151839.
doi: 10.1155/2013/151839 |
| [14] |
LOPEZ A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annual Review of Genetics, 1998, 32:279-305. doi: 10.1146/annurev.genet.32.1.279.
doi: 10.1146/annurev.genet.32.1.279 |
| [15] |
GRAVELEY B R. Alternative splicing: increasing diversity in the proteomic world. Trends in Genetics, 2001, 17(2):100-107. doi: 10.1016/s0168-9525(00)02176-4.
doi: 10.1016/s0168-9525(00)02176-4 |
| [16] |
BARALLE F E, GIUDICE J. Alternative splicing as a regulator of development and tissue identity. Nature Reviews Molecular Cell Biology, 2017, 18(7):437-451. doi: 10.1038/nrm.2017.27.
doi: 10.1038/nrm.2017.27 |
| [17] |
SUNYAEV S, HANKE J, BRETT D, AYDIN A, ZASTROW I, LATHE W, BORK P, REICH J. Individual variation in protein-coding sequences of human genome. Advances in Protein Chemistry, 2000, 54:409-437. doi: 10.1016/s0065-3233(00)54012-1.
doi: 10.1016/s0065-3233(00)54012-1 |
| [18] |
MUROYA S, NAKAJIMA I, CHIKUNI K. Amino acid sequences of multiple fast and slow troponin T isoforms expressed in adult bovine skeletal muscles. Journal of Animal Science, 2003, 81(5):1185-1192. doi: 10.2527/2003.8151185x.
doi: 10.2527/2003.8151185x |
| [19] |
MUROYA S, OHNISHI-KAMEYAMA M, OE M, NAKAJIMA I, CHIKUNI K. Postmortem changes in bovine troponin T isoforms on two-dimensional electrophoretic gel analyzed using mass spectrometry and western blotting: the limited fragmentation into basic polypeptides. Meat Science, 2007, 75(3):506-514. doi: 10.1016/j.meatsci.2006. 08.012.
doi: 10.1016/j.meatsci.2006. 08.012 |
| [20] |
JIN J P, SAMANEZ R A. Evolution of a metal-binding cluster in the NH2-terminal variable region of avian fast skeletal muscle troponin T: functional divergence on the basis of tolerance to structural drifting. Journal of Molecular Evolution, 2001, 52(2):103-116. doi: 10.1007/ s002390010139.
doi: 10.1007/ s002390010139 |
| [21] |
KLEIN P, OLOKO M, ROTH F, MONTEL V, MALERBA A, JARMIN S, GIDARO T, POPPLEWELL L, PERIE S, LACAU ST GUILY J, DE LA GRANGE P, ANTONIOU M N, DICKSON G, BUTLER-BROWNE G, BASTIDE B, MOULY V, TROLLET C. Nuclear poly(A)-binding protein aggregates misplace a pre-mRNA outside of SC35 speckle causing its abnormal splicing. Nucleic Acids Research, 2016. 44(22):10929-10945.
doi: 10.1093/nar/gkw703 |
| [22] |
WEI B, JIN J P. TNNT1, TNNT2, and TNNT3: Isoform genes, regulation, and structure-function relationships. Gene, 2016, 582(1):1-13. doi: 10.1016/j.gene.2016.01.006.
doi: 10.1016/j.gene.2016.01.006 |
| [23] |
SCHIAFFINO S, SANDRI M, MURGIA M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda, Md), 2007, 22:269-278. doi: 10.1152/physiol. 00009.2007.
doi: 10.1152/physiol. 00009.2007 |
| [24] |
LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 |
| [25] |
MEDFORD R M, NGUYEN H T, DESTREE A T, SUMMERS E, NADAL-GINARD B. A novel mechanism of alternative RNA splicing for the developmentally regulated generation of troponin T isoforms from a single gene. Medical Acupuncture, 1984, 38(2):409-421. doi: 10.1016/0092-8674(84)90496-3.
doi: 10.1016/0092-8674(84)90496-3 |
| [26] |
JAYASINGHE R G, CAO S, GAO Q, WENDL M C, VO N S, REYNOLDS S M, ZHAO Y, CLIMENTE-GONZÁLEZ H, CHAI S, WANG F, VARGHESE R, HUANG M, LIANG W W, WYCZALKOWSKI M A, SENGUPTA S, LI Z, PAYNE S H, FENYÖ D, MINER J H, WALTER M J, CANCER GENOME ATLAS RESEARCH NETWORK, VINCENT B, EYRAS E, CHEN K, SHMULEVICH I, CHEN F, DING L. Systematic analysis of splice-site-creating mutations in cancer. Cell Reports, 2018, 23(1): 270-281.e3. doi: 10.1016/j.celrep. 2018.03.052.
doi: 10.1016/j.celrep. 2018.03.052 |
| [27] |
BREITBART R E, NADAL-GINARD B. Complete nucleotide sequence of the fast skeletal troponin T gene. Alternatively spliced exons exhibit unusual inter species divergence. Journal of Molecular Biology, 1986, 188(3):313-324. doi: 10.1016/0022-2836(86)90157-9.
doi: 10.1016/0022-2836(86)90157-9 |
| [28] |
WANG J, JIN J P. Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene, 1997, 193(1):105-114. doi: 10.1016/ s0378-1119(97)00100-5.
doi: 10.1016/ s0378-1119(97)00100-5 |
| [29] |
BRIGGS M M, SCHACHAT F. Origin of fetal troponin T: developmentally regulated splicing of a new exon in the fast troponin T gene. Developmental Biology, 1993, 158(2):503-509. doi: 10.1006/ dbio.1993.1208.
doi: 10.1006/ dbio.1993.1208 |
| [30] |
WU Q L, JHA P K, DU Y, LEAVIS P C, SARKAR S. Overproduction and rapid purification of human fast skeletal beta troponin T using Escherichia coli expression vectors: functional differences between the alpha and beta isoforms. Cancer Biology & Medicine, 1995, 155(2):225-230. doi: 10.1016/0378-1119(94)00846-k.
doi: 10.1016/0378-1119(94)00846-k |
| [31] |
CHALFANT C E, RATHMAN K, PINKERMAN R L, WOOD R E, OBEID L M, OGRETMEN B, HANNUN Y A. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. The Journal of Biological Chemistry, 2002, 277(15):12587-12595. doi: 10.1074/jbc.m112010200.
doi: 10.1074/jbc.m112010200 |
| [32] |
BRANDIMARTI P, COSTA-JÚNIOR J M, FERREIRA S M, PROTZEK A O, SANTOS G J, CARNEIRO E M, BOSCHERO A C, REZENDE L F. Cafeteria diet inhibits insulin clearance by reduced insulin-degrading enzyme expression and mRNA splicing. The Journal of Endocrinology, 2013, 219(2):173-182. doi: 10.1530/joe- 13-0177.
doi: 10.1530/joe- 13-0177 |
| [33] |
MARDEN J H, FESCEMYER H W, SAASTAMOINEN M, MACFARLAND S P, VERA J C, FRILANDER M J, HANSKI I. Weight and nutrition affect pre-mRNA splicing of a muscle gene associated with performance, energetics and life history. Journal of Experimental Biology, 2008. 211(Pt 23):3653-3660.
doi: 10.1242/jeb.023903 |
| [34] |
SCHILDER R J, KIMBALL S R, MARDEN J H, JEFFERSON L S. Body weight-dependent troponin T alternative splicing is evolutionarily conserved from insects to mammals and is partially impaired in skeletal muscle of obese rats. The Journal of Experimental Biology, 2011, 214(pt 9):1523-1532. doi: 10.1242/jeb.051763.
doi: 10.1242/jeb.051763 |
| [35] | BENTZINGER C F, WANG Y X, RUDNICKI M A. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biology, 2012. 4(2). |
| [36] |
CHAL J, POURQUIÉ O. Making muscle: skeletal myogenesis in vivo and in vitro. Development (Cambridge, England), 2017, 144(12):2104-2122. doi: 10.1242/dev.151035.
doi: 10.1242/dev.151035 |
| [37] |
DU M, WANG B, FU X, YANG Q, ZHU M J. Fetal programming in meat production. Meat Science, 2015, 109:40-47. doi: 10.1016/j. meatsci.2015.04.010.
doi: 10.1016/j. meatsci.2015.04.010 |
| [38] |
WEI B, LU Y, JIN J P. Deficiency of slow skeletal muscle troponin T causes atrophy of type I slow fibres and decreases tolerance to fatigue. The Journal of Physiology, 2014, 592(6):1367-1380. doi: 10.1113/ jphysiol.2013.268177.
doi: 10.1113/ jphysiol.2013.268177 |
| [39] | DALY S B, SHAH H, O'SULLIVAN J, ANDERSON B, BHASKAR S, WILLIAMS S, AL-SHEQAIH N, MUEED BIDCHOL A, BANKA S, NEWMAN W G, GIRISHA K M. Exome Sequencing Identifies a Dominant TNNT3 Mutation in a Large Family with Distal Arthrogryposis. Molecular Syndromology, 2014. 5(5):218-228. |
| [40] |
SANDARADURA S A, BOURNAZOS A, MALLAWAARACHCHI A, CUMMINGS B B, WADDELL L B, JONES K J, TROEDSON C, SUDARSANAM A, NASH B M, PETERS G B, ALGAR E M, MACARTHUR D G, NORTH K N, BRAMMAH S, CHARLTON A, LAING N G, WILSON M J, DAVIS M R, COOPER S T. Nemaline myopathy and distal arthrogryposis associated with an autosomal recessive TNNT3 splice variant. Human Mutation, 2018, 39(3):383-388. doi: 10.1002/humu.23385.
doi: 10.1002/humu.23385 |
| [41] |
BIESIADECKI B J, CHONG S M, NOSEK T M, JIN J P. Troponin T core structure and the regulatory NH2-terminal variable region. Biochemistry, 2007, 46(5):1368-1379. doi: 10.1021/bi061949m.
doi: 10.1021/bi061949m |
| [1] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [2] | 李恒,字向东,王会,熊燕,吕明杰,刘宇,蒋旭东. 基于全基因组重测序的山羊产羔数性状关键调控基因的筛选[J]. 中国农业科学, 2022, 55(23): 4753-4768. |
| [3] | 朱春艳,宋佳伟,白天亮,王娜,马帅国,普正菲,董艳,吕建东,李杰,田蓉蓉,罗成科,张银霞,马天利,李培富,田蕾. NaCl胁迫对不同耐盐性粳稻种质幼苗叶绿素荧光特性的影响[J]. 中国农业科学, 2022, 55(13): 2509-2525. |
| [4] | 张卫东,郑玉杰,葛伟,张月朗,李芳,王昕. 单细胞测序对绒山羊毛乳头细胞的鉴定[J]. 中国农业科学, 2022, 55(12): 2436-2446. |
| [5] | 李晓菁,张思雨,刘迪,袁晓伟,李兴盛,石延霞,谢学文,李磊,范腾飞,李宝聚,柴阿丽. 芸薹根肿菌活细胞PMAxx-qPCR快速定量检测方法的建立与应用[J]. 中国农业科学, 2022, 55(10): 1938-1948. |
| [6] | 杜宇,王永,孟庆勇,朱江江,林亚秋. 干扰山羊KLF12促进皮下脂肪细胞分化[J]. 中国农业科学, 2022, 55(1): 184-196. |
| [7] | 许晨,王文静,曹珊,李如雪,张贝贝,孙爱清,张春庆. 花后DA-6处理调控小麦种子活力的机理[J]. 中国农业科学, 2021, 54(9): 1821-1834. |
| [8] | 王涛,韩玉,潘力,王冰,孙茂文,王翌,罗玉子,仇华吉,孙元. 针对非洲猪瘟病毒MGF360-13L基因的TaqMan荧光定量PCR的建立[J]. 中国农业科学, 2021, 54(5): 1073-1080. |
| [9] | 赵乐,杨海丽,李佳璐,杨永恒,张蓉,程文强,成磊,赵永聚. TETs与细胞程序性死亡相关基因在山羊妊娠早期输卵管及子宫角的表达[J]. 中国农业科学, 2021, 54(4): 845-854. |
| [10] | 李天聪,朱行,魏宁,龙凤,武建颖,张燕,董金皋,申珅,郝志敏. 玉米大斑病菌SC35同源基因表达规律与互作分析[J]. 中国农业科学, 2021, 54(4): 733-743. |
| [11] | 冯云奎,王健,马金亮,张柳明,李拥军. miR-31-5p对山羊毛囊干细胞增殖和凋亡的影响[J]. 中国农业科学, 2021, 54(23): 5132-5143. |
| [12] | 赵立群,邱艳红,张晓飞,刘慧,杨静静,张建,张海军,徐秀兰,温常龙. TaqMan探针法实时荧光定量PCR检测西瓜潜隐病毒[J]. 中国农业科学, 2021, 54(20): 4337-4347. |
| [13] | 古明辉,刘永峰,申倩,乔春艳. 猕猴桃多酚在山羊肉冷藏中延缓氧化和改善品质的作用[J]. 中国农业科学, 2020, 53(8): 1643-1654. |
| [14] | 龙凤,王擎,朱行,王建霞,申珅,刘宁,郝志敏,董金皋. 玉米大斑病菌Septin基因家族的鉴定与表达模式分析[J]. 中国农业科学, 2020, 53(24): 5017-5026. |
| [15] | 康俊梅,张俏燕,蒋旭,王珍,张铁军,龙瑞才,崔会婷,杨青川. 紫花苜蓿MsSQE1的克隆及对皂甙合成的功能分析[J]. 中国农业科学, 2020, 53(2): 247-260. |
|
||