













中国农业科学 ›› 2020, Vol. 53 ›› Issue (19): 4092-4102.doi: 10.3864/j.issn.0578-1752.2020.19.021
• 畜牧·兽医·资源昆虫 • 上一篇
收稿日期:2020-01-17
接受日期:2020-03-10
出版日期:2020-10-01
发布日期:2020-10-19
通讯作者:
苏松坤,聂红毅
作者简介:高艳,E-mail: 基金资助:
GAO Yan( ),ZHU YaNan,LI QiuFang,SU SongKun(
),ZHU YaNan,LI QiuFang,SU SongKun( ),NIE HongYi(
),NIE HongYi( )
)
Received:2020-01-17
Accepted:2020-03-10
Online:2020-10-01
Published:2020-10-19
Contact:
SongKun SU,HongYi NIE
摘要:
【目的】意大利蜜蜂(Apis mellifera ligustica)哺育行为在维护蜂群稳定和生产蜂王浆方面发挥重要作用。本研究通过人工组建蜂群获得相同日龄的哺育蜂和采集蜂,筛选出排除日龄因素干扰后哺育蜂脑部与哺育行为密切相关的差异表达基因,揭示哺育蜂脑部调控哺育行为的分子网络。【方法】通过人工组建蜂群的方法获得3日龄工蜂、10日龄哺育蜂和采集蜂、21日龄哺育蜂和采集蜂,解剖各组工蜂头部获得脑组织样本,应用RNA-seq技术对5组脑部样本(3日龄工蜂、10日龄哺育蜂、10日龄采集蜂、21日龄哺育蜂、21日龄采集蜂)中基因表达量进行转录组测序的全面分析,筛选出哺育蜂脑部哺育行为密切相关的差异表达基因,并对这些差异表达基因进行GO和KEGG富集分析。同时利用实时荧光定量PCR(qPCR)对随机选取的4个差异表达基因的表达模式进行验证。【结果】RNA-seq分析筛选得到32个与哺育蜂哺育行为密切相关的差异表达基因,这些基因在10日龄哺育蜂脑中表达量均显著高于3日龄工蜂、10日龄采集蜂、21日龄采集蜂,且在21日龄哺育蜂脑中的表达量也显著高于21日龄采集蜂。GO富集分析发现上调差异表达基因主要参与氧化还原酶活性、气味结合、跨膜运输等功能。KEGG富集结果显示,上调的差异表达基因主要参与蛋白质代谢(核糖体)、能量代谢(氧化磷酸化、碳代谢、三羧酸循环、淀粉和蔗糖代谢、氮代谢、其他聚糖降解通路)、信号转导(Toll和Imd信号通路、光传导、鞘脂代谢)、消化作用(溶酶体),其中显著性富集在鞘脂代谢和其他聚糖降解通路。qPCR结果显示3个上调差异表达基因(LOC409709、LOC551813、LOC409708)和1个下调差异表达基因(LOC551165)表达模式的检测结果与测序数据一致。【结论】通过对3日龄工蜂、10日龄哺育蜂、10日龄采集蜂、21日龄哺育蜂、21日龄采集蜂5组样本脑部进行全面分析,获得相同日龄哺育蜂和采集蜂脑部基因的转录组图谱,分析得到了脑部哺育行为相关的32个上调差异表达基因。哺育蜂脑部的差异表达基因主要通过信号转导和能量代谢等途径调控哺育蜂的哺育行为。
高艳,朱雅楠,李秋方,苏松坤,聂红毅. 转录组学分析意大利蜜蜂脑部哺育行为相关基因[J]. 中国农业科学, 2020, 53(19): 4092-4102.
GAO Yan,ZHU YaNan,LI QiuFang,SU SongKun,NIE HongYi. Transcriptomic Analysis of Genes Related to Nursing Behavior in the Brains of Apis mellifera ligustica[J]. Scientia Agricultura Sinica, 2020, 53(19): 4092-4102.
表1
qPCR引物信息"
| 基因 Gene | 登录号 Accession number | 引物序列 Primer sequence (5′-3′) | 产物大小 Amplicon length (bp) |
|---|---|---|---|
| LOC409709 | 409709 | F: TTGGTCAGTTGGATGGGTAGA | 236 |
| R: CCGACGGTGTAAGAAAGGC | |||
| LOC551813 | 551813 | F: CAAAATTCAACAGCTCCTGC | 210 |
| R: TCATCGCTACCGAAATCATAA | |||
| LOC409708 | 409708 | F: TTGTTGATCGCAATCCTGTC | 480 |
| R: CGTCGCATCGTCATCGTAA | |||
| LOC551165 | 551165 | F:TGCCTGTTGTATTCTCGTAT | 304 |
| R:TCTGACCTTGCCCTCCTC | |||
| Actin | NM_001185146.1 | F: CCTAGCACCATCCACCATGAA | 87 |
| R: GAAGCAAGAATTGACCCACCAA |
表2
RNA-seq数据总览"
| 样品 Sample | 原始读数 Number of raw reads | 有效读数 Number of clean reads | 有效匹配数(匹配率) Clean map (Map rate) | 单一匹配数(匹配率)Unique map (Map rate) | 多匹配数(匹配率)Multiple map (Map rate) | 99.9%的碱基正确率99.9% base accuracy (%) |
|---|---|---|---|---|---|---|
| 3 d_1 | 67515798 | 66708824 | 56766708 (85.10%) | 55951632 (83.87%) | 815076 (1.22%) | 88.25 |
| 3 d_2 | 83920302 | 81832028 | 70899666 (86.64%) | 69821133 (85.32%) | 1078533 (1.32%) | 90.59 |
| 3 d_3 | 81546254 | 80442702 | 69502321 (86.40%) | 68471536 (85.12%) | 1030785 (1.28%) | 89.66 |
| 10 dN_1 | 57288572 | 56348822 | 49321933 (87.53%) | 48015717 (85.21%) | 1306216 (2.32%) | 90.51 |
| 10 dN_2 | 83787622 | 82964416 | 63408429 (76.43%) | 60124557 (72.47%) | 3283872 (3.96%) | 90.66 |
| 10 dN_3 | 72837334 | 71663646 | 56922446 (79.43%) | 56028890 (78.18%) | 893556 (1.25%) | 91.00 |
| 10 dF_1 | 67610626 | 66671128 | 55057317 (82.58%) | 54208585 (81.31%) | 848732 (1.27%) | 90.37 |
| 10 dF_2 | 81258562 | 80246018 | 63725456 (79.41%) | 62620480 (78.04%) | 1104976 (1.38%) | 90.06 |
| 10 dF_3 | 66021926 | 65365544 | 52522944 (80.35%) | 51744723 (79.16%) | 778221 (1.19%) | 91.00 |
| 21 dN_1 | 71882296 | 71020506 | 56181047 (79.11%) | 54846870 (77.23%) | 8223024 (13.18%) | 92.19 |
| 21 dN_2 | 72018192 | 70803290 | 60295752 (85.16%) | 58523770 (82.66%) | 11236986 (15.51%) | 92.33 |
| 21 dN_3 | 72458810 | 71460376 | 59684702 (83.52%) | 57902404 (81.03%) | 8742852 (13.63%) | 92.50 |
| 21 dF_1 | 72168956 | 70890254 | 59385891 (83.77%) | 58485427 (82.50%) | 900464 (1.27%) | 91.94 |
| 21 dF_2 | 69418794 | 68335020 | 55559809 (81.31%) | 54623413 (79.93%) | 936396 (1.37%) | 88.98 |
| 21 dF_3 | 81670548 | 80692702 | 67432186 (83.57%) | 66368993 (82.25%) | 1063193 (1.32%) | 91.86 |
表3
脑部41个DEG基于日龄依赖性在5组样本中的显著性趋势"
| 基因数目 Gene number | 10 dN vs 3 d显著性比较Significant comparison of 10 dN vs 3 d | 10 dN vs 21 dF显著性比较Significant comparison of 10 dN vs 21 dF | 10 dN vs 10 dF显著性比较Significant comparison of 10 dN vs 10 dF | 21 dN vs 21 dF显著性比较Significant comparison of 21 dN vs 21 dF | 10 dN vs 21 dN显著性比较 Significant comparison of 10 dN vs 21 dN |
|---|---|---|---|---|---|
| 32 | ↑ | ↑ | ↑ | ↑ | ns |
| 6 | ↓ | ↓ | ↓ | ↓ | ns |
| 3 | ↑ | ↓ | ↓ | ↓ | ns |
表4
脑部前11个上调DEG在5组样本中的表达信息"
| 基因登录号 Gene_ID | 基因 Gene | 基因描述 Gene description | 平均FPKM值 Average FPKM value | log2 FC (10 dN vs 10 dF) | log2 FC (21 dN vs 21 dF) | ||||
|---|---|---|---|---|---|---|---|---|---|
| 3 d | 10 dN | 10 dF | 21 dN | 21 dF | |||||
| 551813 | LOC551813 | 王浆主蛋白1 Major royal jelly protein 1 | 0.40 | 1564.78 | 2.27 | 510.71 | 9.64 | 9.44 | 5.71 |
| 725215 | LOC725215 | 毒酸磷酸酶Acph-1样 Venom acid phosphatase Acph-1-like | 0.31 | 29.97 | 0.66 | 23.80 | 0.73 | 5.52 | 5.01 |
| 411186 | LOC411186 | 突触小泡糖蛋白2C Synaptic vesicle glycoprotein 2C | 0.22 | 12.81 | 0.47 | 11.96 | 0.60 | 4.78 | 4.29 |
| 410149 | LOC410149 | 羧肽酶Q样 Carboxypeptidase Q-like | 1.59 | 31.09 | 1.82 | 26.29 | 2.81 | 4.10 | 3.21 |
| 409709 | LOC409709 | 假定的葡萄糖基神经酰胺酶4 Putative glucosylceramidase 4 | 2.83 | 75.03 | 7.81 | 50.90 | 8.63 | 3.27 | 2.54 |
| 552478 | Obp17 | 气味结合蛋白17 Odorant binding protein 17 | 1.77 | 6.43 | 0.73 | 4.05 | 0.80 | 3.14 | 2.33 |
| 551935 | Obp21 | 气味结合蛋白21 Odorant binding protein 21 | 4.50 | 34.70 | 4.43 | 22.20 | 7.71 | 2.97 | 1.51 |
| 677673 | Obp14 | 气味结合蛋白14 Odorant binding protein 14 | 1.00 | 7.70 | 1.01 | 3.32 | 0.69 | 2.92 | 2.24 |
| 409708 | LOC409708 | 葡糖基神经酰胺酶样 Glucosylceramidase-like | 13.84 | 132.92 | 22.42 | 113.84 | 27.93 | 2.57 | 2.01 |
| 726798 | LOC726798 | Sn1特异性二酰基甘油脂肪酶β Sn1-specific diacylglycerol lipase beta | 0.53 | 2.67 | 0.49 | 3.46 | 0.85 | 2.45 | 2.01 |
| 408383 | CYP6AQ1 | 细胞色素P450 6AQ1 Cytochrome P450 6AQ1 | 0.49 | 1.97 | 0.42 | 1.41 | 0.39 | 2.22 | 1.84 |
表5
上调DEG富集的GO条目"
| GO条目 GO term | 基因 Gene |
|---|---|
| 氧化还原酶活性 Oxidoreductase activity | CYP6AQ1/LOC726418/ LOC408734/LOC413590 |
| 气味结合 Odorant binding | Obp21/Obp14/Obp17 |
| 跨膜运输 Transmembrane transport | LOC411186/LOC725462 |
| 蛋白质水解 Proteolysis | LOC725215/LOC725154 |
| 翻译 Translation | LOC550715 |
| 脂质代谢 Lipid metabolism | LOC726798 |
| DNA复制 DNA replication | LOC725238 |
| 细胞器组分Organelle part | LOC408444 |
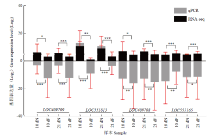
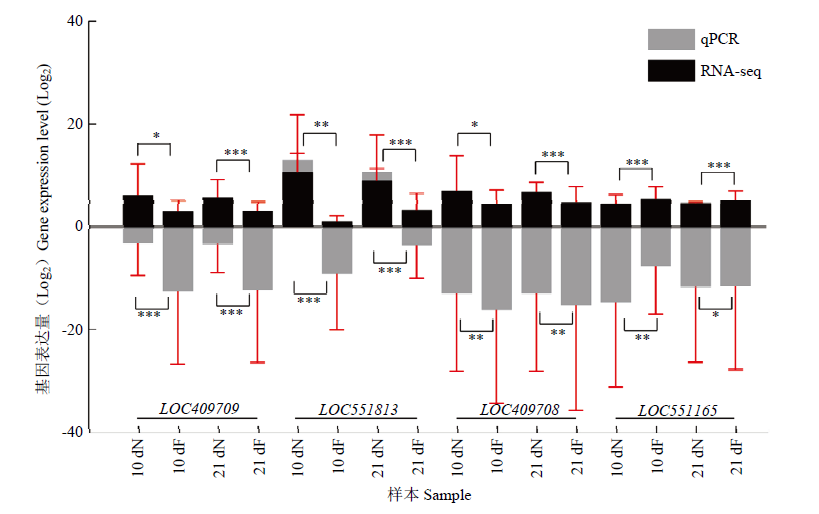
图3
转录组数据的qPCR验证 黑色柱状图表示RNA-seq数据中基因的FPKM值,灰色表示qPCR数据中基因的相对表达量,纵坐标均为相应取过以2为底的对数值The black bar graph represents the FPKM value of the gene in the RNA-seq data, and the gray represents the relative expression of genes in the qPCR data; the ordinates are the corresponding logarithmic values with a base of 2。图中数据为平均值±标准误,采用单因素方差分析 Data in the figure are the mean±SE, One-way ANOVA. *P<0.05; **P<0.01; ***P<0.001"
| [1] |
BEZABIH G, CHENG H, HAN B, FENG M, XUE Y, HU H, LI J K. Phosphoproteome analysis reveals phosphorylation underpinnings in the brains of nurse and forager honeybees ( Apis mellifera). Scientific Reports, 2017, 7: 1973.
doi: 10.1038/s41598-017-02192-3 pmid: 28512345 |
| [2] |
HAN B, FANG Y, FENG M, HU H, HAO Y, MA C, HUO X, MENG L, ZHANG X, WU F, LI J K. Brain membrane proteome and phosphoproteome reveal molecular basis associating with nursing and foraging behaviors of honeybee workers. Journal of Proteome Research, 2017, 16(10): 3646-3663.
doi: 10.1021/acs.jproteome.7b00371 pmid: 28879772 |
| [3] |
SIALANA F J, MENEGASSO A R S, SMIDAK R, HUSSEIN A M, ZAVADIL M, RATTEI T, LUBEC G, PALMA M S, LUBEC J. Proteome changes paralleling the olfactory conditioning in the forager honey bee and provision of a brain proteomicsdataset. Proteomics, 2019, 19(13): e1900094.
doi: 10.1002/pmic.201900094 pmid: 31115157 |
| [4] |
CONTE Y L, MOHAMMEDI A, ROBINSON G E. Primer effects of a brood pheromone on honeybee behavioural development. Proceedings. Biological Sciences, 2001, 268(1463): 163-168.
doi: 10.1098/rspb.2000.1345 pmid: 11209886 |
| [5] |
FUJITA T, KOZUKA-HATA H, AO-KONDO H, KUNIEDA T, OYAMA M, KUBO T. Proteomic analysis of the royal jelly and characterization of the functions of its derivation glands in the honeybee. Journal of Proteome Research, 2012, 12(1): 404-411.
doi: 10.1021/pr300700e pmid: 23157659 |
| [6] |
ALTAYE S Z, MENG L F, LI J K. Molecular insights into the enhanced performance of royal jelly secretion by a stock of honeybee ( Apis mellifera ligustica) selected for increasing royal jelly production. Apidologie, 2019, 50(4): 436-453.
doi: 10.1007/s13592-019-00656-1 |
| [7] |
WHITFIELD C W, CZIKO A M, ROBINSON G E. Gene expression profiles in the brain predict behavior in individual honey bees. Science, 2003, 302(5643): 296-299.
doi: 10.1126/science.1086807 pmid: 14551438 |
| [8] |
KUCHARSKI R, MALESZKA R. Evaluation of differential gene expression during behavioral development in the honeybee using microarrays and northern blots. Genome Biology, 2002, 3(2): RESEARCH0007.
doi: 10.1186/gb-2002-3-2-research0007 pmid: 11864369 |
| [9] |
RODRIGUEZ-ZAS S L, SOUTHEY B R, SHEMESH Y, RUBIN E B, COHEN M, ROBINSON G E, BLOCH G. Microarray analysis of natural socially regulated plasticity in circadian rhythms of honey bees. Journal of Biological Rhythms, 2012, 27(1): 12-24.
doi: 10.1177/0748730411431404 pmid: 22306970 |
| [10] |
HERNÁNDEZ L G, LU B, DA CRUZ G C, CALÁBRIA L K, MARTINS N F, TOGAWA R, ESPINDOLA F S, YATES J R, CUNHA R B, DE SOUSA M V. Worker honeybee brain proteome. Journal of Proteome Research, 2012, 11(3): 1485-1493.
doi: 10.1021/pr2007818 pmid: 22181811 |
| [11] |
PAERHATI Y, ISHIGURO S, UEDA-MATSUO R, YANG P, YAMASHITA T, ITO K, MAEKAWA H, TANI H, SUZUKI K. Expression of AmGR10 of the gustatory receptor family in honey bee is correlated with nursing behavior. PLoS ONE, 2015, 10(11): e0142917.
doi: 10.1371/journal.pone.0142917 pmid: 26588091 |
| [12] |
KNECHT D, KAATZ H H. Patterns of larval food production by hypopharyngeal glands in adult worker honey bees. Apidologie, 1990, 21(5): 457-468.
doi: 10.1051/apido:19900507 |
| [13] |
HAN B, FANG Y, FENG M, HU H, QI Y P, HUO X M, MENG L F, WU B, LI J K. Quantitative neuropeptidome analysis reveals neuropeptides are correlated with social behavior regulation of the honeybee workers. Journal of Proteome Research, 2015, 14(10): 4382-4393.
pmid: 26310634 |
| [14] | 韩宾. 工蜂劳动分工与蜂王浆高产机理的大脑神经肽组、膜蛋白质组和膜磷酸化蛋白质组研究[D]. 北京: 中国农业科学院, 2017. |
| HAN B. Investigation of molecular basis associating with division of labor and high royal jelly yields by analyzing brain neuropeptidome, membrane proteome and membrane phosphoproteome[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. (in Chinese) | |
| [15] |
TSUCHIMOTO M, AOKI M, TAKADA M, KANOU Y, SASAGAWA H, KITAGAWA Y, KADOWAKI T. The changes of gene expression in honeybee ( Apis mellifera) brains associated with ages. Zoological Science, 2004, 21(1): 23-28.
pmid: 14745100 |
| [16] | 赵元洪, 赵晓蒙, 苏松坤. 蜜蜂全脑解剖新方法的研究. 中国蜂业, 2014, 65(Z1): 4-7. |
| ZHAO Y H, ZHAO X M, SU S K. New method on dissection of whole brain of honeybee. Apiculture of China, 2014, 65(Z1): 4-7. (in Chinese) | |
| [17] |
ANDERS S, HUBER W. Differential expression analysis for sequence count data. Genome Biology, 2010, 11(10): R106.
doi: 10.1186/gb-2010-11-10-r106 pmid: 20979621 |
| [18] |
LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [19] |
SCHULZ D J, BARRON A B, ROBINSON G E. A role for octopamine in honey bee division of labor. Brain, Behavior and Evolution, 2002, 60(6): 350-359.
doi: 10.1159/000067788 pmid: 12563167 |
| [20] |
XU G, TENG Z W, GU G X, QI Y X, GUO L, XIAO S, WANG F, FANG Q, WANG F, SONG Q S, STANLEY D, YE G Y. Genome- wide characterization and transcriptomic analyses of neuropeptides and their receptors in an endoparasitoid wasp, Pteromalus puparum. Archives of Insect Biochemistry and Physiology, 2019, 103(2): e21625.
pmid: 31565815 |
| [21] |
PRATAVIEIRA M, DA SLIVA MENEGASSO A R, ESTEVES F G, SATO K U, MALASPINA O, PALMA M S. MALDI imaging analysis of neuropeptides in africanized honeybee ( Apis mellifera) brain: Effect of aggressiveness. Journal of Proteome Research, 2018, 17(7): 2358-2369.
doi: 10.1021/acs.jproteome.8b00098 pmid: 29775065 |
| [22] |
LIU Z, JI T, YIN L, SHEN J, SHEN F, CHEN G. Transcriptome sequencing analysis reveals the regulation of the hypopharyngeal glands in the honey bee, Apis mellifera carnica Pollmann. PLoS ONE, 2013, 8(12): e81001.
doi: 10.1371/journal.pone.0081001 pmid: 24339892 |
| [23] |
WU Y Q, ZHENG H Q, CORONA M, PIRK C, MENG F, ZHENG Y F, HU F L. Comparative transcriptome analysis on the synthesis pathway of honey bee ( Apis mellifera) mandibular gland secretions. Scientific Reports, 2017, 7(1): 4530.
doi: 10.1038/s41598-017-04879-z pmid: 28674395 |
| [24] |
STOUT K A, DUNN A R, HOFFMAN C, MILLER G W. The synaptic vesicle glycoprotein 2: Structure, function, and disease relevance. ACS Chemical Neuroscience, 2019, 10(9): 3927-3938.
pmid: 31394034 |
| [25] |
MAZZUCCHELLI C, BRAMBILLA R. Ras-related and MAPK signalling in neuronal plasticity and memory formation. Cellular and Molecular Life Sciences, 2000, 57(4): 604-611.
doi: 10.1007/PL00000722 pmid: 11130460 |
| [26] |
ASTUDILLO L, THERVILLE N, COLACIOS C, SÉGUI B, ANDRIEU-ABADIE N, LEVADE T . Glucosylceramidases and malignancies in mammals. Biochimie, 2016, 125: 267-280.
doi: 10.1016/j.biochi.2015.11.009 pmid: 26582417 |
| [27] | 孙九丽, 林慧珍, 苟萍. 鞘脂代谢及其相关疾病研究进展. 生物技术, 2011, 21(5): 93-97. |
| SUN J L, LIN H Z, GOU P. Research progress of sphingolipid metabolism and related diseases. Biotechnology, 2011, 21(5): 93-97. (in Chinese) | |
| [28] |
SHI J, ANDERSON D, LYNCH B A, CASTAIGNE J G, FOERCH P, LEBON F. Combining modelling and mutagenesis studies of synaptic vesicle protein 2A to identify a series of residues involved in racetam binding. Biochemical Society Transactions, 2011, 39(5): 1341-1347.
doi: 10.1042/BST0391341 pmid: 21936812 |
| [29] |
BISOGNO T, HOWELL F, WILLIAMS G, MINASSI A, CASCIO M, LIGRESTI A, MATIAS I, SCHIANO-MORIELLO A, PAUL P, WILLIAMS E J, GANGADHARAN U, HOBBS C, DI MARZO V, DOHERTY P. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. The Journal of Cell Biology, 2003, 163(3): 463-468.
pmid: 14610053 |
| [30] |
BRITTIS P A, SILVER J, WALSH F S, DOHERTY P. Fibroblast growth factor receptor function is required for the orderly projection of ganglion cell axons in the developing mammalian retina. Molecular and Cellular Neuroscience, 1996, 8(2/3): 120-128.
doi: 10.1006/mcne.1996.0051 pmid: 8954627 |
| [31] |
RIDGE K D, ABDULAEV N G, SOUSA M, PALCZEWSKI K. Phototransduction: Crystal clear. Trends in Biochemical Sciences, 2003, 28(9): 479-487.
doi: 10.1016/S0968-0004(03)00172-5 pmid: 13678959 |
| [32] |
STEINBRECHT R A. Odorant-binding proteins: Expression and function. Annals of the New York Academy of Sciences, 1998, 855(1): 323-332.
doi: 10.1111/nyas.1998.855.issue-1 |
| [33] | BRIAND L, SWASDIPAN N, NESPOULOUS C, BÉZIRARD V, BLON F, HUET J C, EBERT P, PERNOLLET J C. Characterization of a chemosensory protein (ASP3c) from honeybee ( Apis mellifera L.) as a brood pheromone carrier. The FEBS Journal, 2002, 269(18): 4586-4596. |
| [34] |
IOVINELLA I, DANI F, NICCOLINI A, SIMONA S, MICHELUCCI E, GAZZANO A, TURILLAZZI S, FELICIOLI A, PELOSI P. Differential expression of odorant-binding proteins in the mandibular glands of the honey bee according to caste and age. Journal of Proteome Research, 2011, 10(8): 3439-3449.
doi: 10.1021/pr2000754 pmid: 21707107 |
| [35] |
WU F, FENG Y L, HAN B, HU H, FENG M, MENG L F, MA C, YU L S, LI J K. Mechanistic insight into binding interaction between chemosensory protein 4 and volatile larval pheromones in honeybees ( Apis mellifera). International Journal of Biological Macromolecules, 2019, 141: 553-563.
doi: 10.1016/j.ijbiomac.2019.09.041 pmid: 31499112 |
| [36] |
NIE H Y, XU S P, XIE C Q, GENG H Y, ZHAO Y Z, LI J H, HUANG W F, LIN Y, LI Z G, SU S K. Comparative transcriptome analysis of Apis mellifera antennae of workers performing different tasks. Molecular Genetics and Genomics, 2017, 293(1): 237-248.
doi: 10.1007/s00438-017-1382-5 pmid: 29043489 |
| [37] |
LESNEFSKY E J, HOPPEL C L. Oxidative phosphorylation and aging. Ageing Research Reviews, 2006, 5(4): 402-433.
doi: 10.1016/j.arr.2006.04.001 pmid: 16831573 |
| [38] | 唐晓伟. 西方蜜蜂细胞色素P450单加氧酶特性初步研究[D]. 北京: 中国农业科学院, 2011. |
| TANG X W. Preliminary study on cytochrome P450 monooxygenase of Apis mellifera[D]. Beijing: Chinese Academy of Agricultural Sciences, 2011. (in Chinese) | |
| [39] |
HELVIG C, TIJET N, FEYEREISEN R, WALKER F A, RESTIFO L L. Drosophila melanogaster CYP6A8, an insect P450 that catalyzes lauric acid (omega-1)-hydroxylation. Biochemical and Biophysical Research Communications, 2004, 325(4): 1495-1502.
doi: 10.1016/j.bbrc.2004.10.194 pmid: 15555597 |
| [40] |
BOUTIN S, ALBURAK M, MERCIER P L, GIOVENAZZO P, DEROME N. Differential gene expression between hygienic and non-hygienic honeybee ( Apis mellifera L.) hives. BMC Genomics, 2015, 16(1): 500.
doi: 10.1186/s12864-015-1714-y |
| [41] |
GARCIA L, GARCIA C H, CALÁBRIA L K, DA CRUZ G C, PUENTES A S, BÁO S N, FONTES W, RICART C A, ESPINDOLA F S, DE SOUSA M V. Proteomic analysis of honey bee brain upon ontogenetic and behavioral development. Journal of Proteome Research, 2009, 8(3): 1464-1473.
pmid: 19203288 |
| [1] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [2] | 张晓萍,撒世娟,伍涵宇,乔丽媛,郑蕊,姚新灵. 马铃薯叶片气孔的开张与关闭同步伴随果胶的降解与合成[J]. 中国农业科学, 2022, 55(17): 3278-3288. |
| [3] | 王荣华,孟丽峰,冯毛,房宇,魏俏红,马贝贝,钟未来,李建科. 蜂王浆高产蜜蜂与意大利蜜蜂哺育蜂唾液腺蛋白质组分析[J]. 中国农业科学, 2022, 55(13): 2667-2684. |
| [4] | 杜宇,范小雪,蒋海宾,王杰,冯睿蓉,张文德,余岢骏,隆琦,蔡宗兵,熊翠玲,郑燕珍,陈大福,付中民,徐国钧,郭睿. 微小RNA介导意大利蜜蜂工蜂对东方蜜蜂微孢子虫的跨界调控[J]. 中国农业科学, 2021, 54(8): 1805-1820. |
| [5] | 朱芳芳,董亚辉,任真真,王志勇,苏慧慧,库丽霞,陈彦惠. 过表达ZmIBH1-1提高玉米苗期抗旱性[J]. 中国农业科学, 2021, 54(21): 4500-4513. |
| [6] | 刘锴,何闪闪,张彩霞,张利义,卞书迅,袁高鹏,李武兴,康立群,丛佩华,韩晓蕾. 苹果叶片不定芽再生过程的差异表达基因鉴定与分析[J]. 中国农业科学, 2021, 54(16): 3488-3501. |
| [7] | 张稳,孟淑君,王琪月,万炯,马拴红,林源,丁冬,汤继华. 玉米pTAC2影响苗期叶片叶绿素合成的转录组分析[J]. 中国农业科学, 2020, 53(5): 874-889. |
| [8] | 徐志军,赵胜,徐磊,胡小文,安东升,刘洋. 基于RNA-seq数据的栽培种花生SSR位点鉴定和标记开发[J]. 中国农业科学, 2020, 53(4): 695-706. |
| [9] | 张丽翠,马川,冯毛,李建科. 基于高分辨质谱和代谢组学技术评估和优化蜂王浆代谢物提取方法[J]. 中国农业科学, 2020, 53(18): 3833-3845. |
| [10] | 耿四海,石彩云,范小雪,王杰,祝智威,蒋海宾,范元婵,陈华枝,杜宇,王心蕊,熊翠玲,郑燕珍,付中民,陈大福,郭睿. 微小RNA介导东方蜜蜂微孢子虫侵染意大利蜜蜂工蜂的分子机制[J]. 中国农业科学, 2020, 53(15): 3187-3204. |
| [11] | 杜宇,范小雪,蒋海宾,王杰,范元婵,祝智威,周丁丁,万洁琦,卢家轩,熊翠玲,郑燕珍,陈大福,郭睿. 微小RNA及其介导的竞争性内源RNA调控网络在意大利蜜蜂工蜂中肠发育过程中的潜在作用[J]. 中国农业科学, 2020, 53(12): 2512-2526. |
| [12] | 杜宇,周丁丁,万洁琦,卢家轩,范小雪,范元婵,陈恒,熊翠玲,郑燕珍,付中民,徐国钧,陈大福,郭睿. 意大利蜜蜂工蜂中肠发育过程中的差异基因 表达谱及调控网络[J]. 中国农业科学, 2020, 53(1): 201-212. |
| [13] | 郭书磊,鲁晓民,齐建双,魏良明,张新,韩小花,岳润清,王振华,铁双贵,陈彦惠. 利用RNA-Seq发掘玉米叶片形态建成相关的调控基因[J]. 中国农业科学, 2020, 53(1): 1-17. |
| [14] | 梁国平,李文芳,陈佰鸿,左存武,马丽娟,何红红,万鹏,安泽山,毛娟. 不同糖源对葡萄试管苗蛋白激酶相关基因表达的影响[J]. 中国农业科学, 2019, 52(7): 1119-1135. |
| [15] | 金梅,张丽娟,曹倩,郭鑫英. MT和FGF5调控辽宁绒山羊绒毛生长相关LncRNA的筛选及鉴定[J]. 中国农业科学, 2019, 52(4): 738-754. |
|
||