













中国农业科学 ›› 2019, Vol. 52 ›› Issue (3): 503-511.doi: 10.3864/j.issn.0578-1752.2019.03.010
收稿日期:2018-08-27
接受日期:2018-09-28
出版日期:2019-02-01
发布日期:2019-02-14
作者简介:宋杨,E-mail: 基金资助:
SONG Yang,LIU HongDi,WANG HaiBo,ZHANG HongJun,LIU FengZhi( )
)
Received:2018-08-27
Accepted:2018-09-28
Online:2019-02-01
Published:2019-02-14
Supported by:摘要:
目的 分离越橘VcNAC072(NAM,ATAF1/2,CUC2)转录因子,分析其表达模式并探讨其在调控花青素合成过程中的功能,为进一步研究越橘花青素积累的调控机理提供理论基础。方法 以‘公爵’越橘(Vaccinium corymbosum ‘Duke’)为试材,克隆VcNAC072。通过农杆菌介导法获得转基因拟南芥,比较转基因和野生型拟南芥花青素积累的差异。利用酵母单杂交和瞬时表达试验,分析VcNAC072对MYB转录因子AtPAP1的转录调控。结果 克隆获得越橘VcNAC072,该基因CDS为1 032 bp,编码含有343个氨基酸的蛋白质,含有1个保守的NAC结构域。表达分析显示,该基因在不同发育阶段的果实中均可表达,但表达差异明显,在粉色和蓝色果实中表达量较高,在绿色果实中表达量最低。随着VcNAC072表达的升高,果实中花青素含量呈递增的趋势。分析AtPAP1启动子序列,发现其序列中包含NAC转录因子的结合位点。酵母单杂交和烟草瞬时表达试验结果表明,VcNAC072可与AtPAP1的启动子相互作用,并激活其表达。在野生型拟南芥中异位表达VcNAC072,其种子中花青素积累量显著高于野生型。结论 推测VcNAC072在越橘果实中正向调节花青素的积累。
宋杨,刘红弟,王海波,张红军,刘凤之. 越橘VcNAC072克隆及其促进花青素积累的功能分析[J]. 中国农业科学, 2019, 52(3): 503-511.
SONG Yang,LIU HongDi,WANG HaiBo,ZHANG HongJun,LIU FengZhi. Molecular Cloning and Functional Characterization of VcNAC072 Reveals Its Involvement in Anthocyanin Accumulation in Blueberry[J]. Scientia Agricultura Sinica, 2019, 52(3): 503-511.
表1
本研究中使用的引物"
| 用途 Use | 引物名称Primer name | 序列(5′-3′)Primer sequence (5′-3′) |
|---|---|---|
| 构建pRI101-VcNAC072表达载体 The construction of pRI101-VcNAC072 expression vector 构建pGADT7-VcNAC072表达载体 The construction of pGADT7-VcNAC072 expression vector 构建pAbAi-AtPAP1pro表达载体 The construction of pAbAi-AtPAP1pro expression vector 构建AtPAP1pro-GUS表达载体 The construction of AtPAP1pro-GUS expression vector 实时荧光定量qRT-PCR Real time fluorescent quantitative PCR | VcNAC072(pRI101)-F VcNAC072(pRI101)-R VcNAC072( pGADT7)-F VcNAC072( pGADT7)-R AtPAP1pro(pAbAi)-F AtPAP1pro(pAbAi)-R AtPAP1pro(GUS)-F AtPAP1pro(GUS)-R VcNAC072-(RT-PCR)-F VcNAC072-(RT-PCR)-R VcGAPDH-(RT-PCR)-F VcGAPDH-(RT-PCR)-R AtUBQ10-(RT-PCR)-F AtUBQ10-(RT-PCR)-R AtPAP1-(RT-PCR)-F AtPAP1-(RT-PCR)-R AtDFR-(RT-PCR)-F AtDFR-(RT-PCR)-R AtANS-(RT-PCR)-F AtANS-(RT-PCR)-R | GGAATTCCATATGATGGGAGTTCAGGAGTCCG CGCGGATCCTCACTGCCGAAACCCGAATCC GGAATTCCATATGATGGGAGTTCAGGAGTCCG CGCGGATCCTCACTGCCGAAACCCGAATCC CGAGCTCCTAATACATAAAATGTGGATATC ACGCGTCGACGGAACAAAGATAGATACGTAA CGGGGTACCCTAATACATAAAATGTGGATATC CATGCCATGGGGAACAAAGATAGATACGTAA GATGAGGTACAATGACATG GAGTTCTGCTGAAACAAC GGTTATCAATGATAGGTTTGGCA CAGTCCTTGCTTGATGGACC CGTTAAGACGTTGACTGGGAAAACT GCTTTCACGTTATCAATGGTGTCA GCTCTGATGAAGTCGATCTTC CTACCTCTTGGCTTTCCTCT GTCGGTCCATTCATCACAAC TGAGCGTTGCATAAGTCGTC TCAAGAAAGCCGGAGAAGAG TTGTCCACTCGCGTTGTTAG |

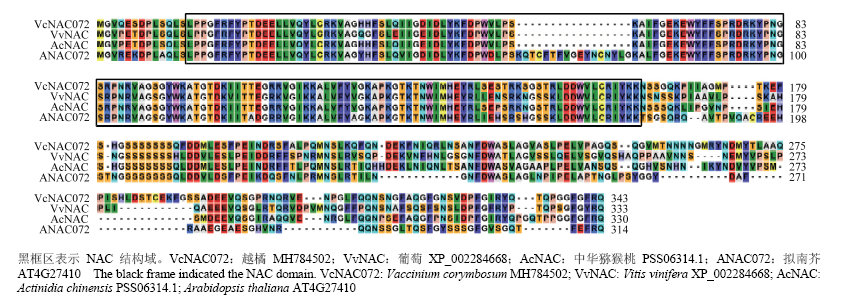
图1
VcNAC072与其他植物NAC蛋白的多序列比较黑框区表示NAC结构域。VcNAC072:越橘 MH784502;VvNAC:葡萄 XP_002284668;AcNAC:中华猕猴桃 PSS06314.1;ANAC072:拟南芥 AT4G27410 The black frame indicated the NAC domain. VcNAC072: Vaccinium corymbosum MH784502; VvNAC: Vitis vinifera XP_002284668; AcNAC: Actinidia chinensis PSS06314.1; Arabidopsis thaliana AT4G27410"
| [1] | 李亚东, 孙海悦, 陈丽 . 我国蓝莓产业发展报告.中国果树, 2016(5):1-10. |
| LI Y D, SUN H Y, CHEN L . Report on the development of blueberry industry in China. China Fruits, 2016(5):1-10. (in Chinese) | |
| [2] | 吴林 . 中国蓝莓35年—科学研究与产业发展. 吉林农业大学学报, 2016,38(1):1-11. |
| WU L . Thirty-five years of research and industry development of blueberry in China. Journal of Jilin Agricultural University, 2016,38(1):1-11. (in Chinese) | |
| [3] |
GORDILLO G, FANG H Q, KHANNA S, HARPER J, PHILIPS G, SEN C K . Oral administration of blueberry inhibits angiogenic tumor growth and enhances survival of mice with endothelial cell neoplasm. Antioxid Redox Signal, 2009,11(1):47-58.
doi: 10.1089/ars.2008.2150 pmid: 2933151 |
| [4] |
BASU A, RHONE M, LYONS T J . Berries: Emerging impact on cardiovascular health. Nutrition Reviews, 2010,68(3):168-177.
doi: 10.1111/j.1753-4887.2010.00273.x pmid: 20384847 |
| [5] | SHAHNEJAT-BUSHEHRI S, TARKOWSKA D, SAKURABA Y, BALAZADEH S . Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nature Plants, 2016,2:16013. |
| [6] | KIM H S, PARK B O, YOO J H, JUNG M S, LEE S M, HAN H J, KIM K E, KIM S H, LIM C O, YUN D J, LEE S Y, CHUNG W S . Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. Journal of Biological Chemistry, 2007,282(50):36292-36302. |
| [7] |
HONG Y B, ZHANG H J, HUANG L, LI D Y, SONG F M . Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Frontiers in Plant Science, 2016,7(e0116646):4.
doi: 10.3389/fpls.2016.00004 pmid: 26834774 |
| [8] |
YU X W, LIU Y M, WANG S, TAO Y, WANG Z K, MIJITI A, WANG Z, ZHANG H, MA H . A chickpea stress-responsive NAC transcription factor,CarNAC5, confers enhanced tolerance to drought stress in transgenic. Plant Growth Regulation, 2016,79(2):187-197.
doi: 10.1007/s10725-015-0124-0 |
| [9] |
KO J H, YANG S H, PARK A H, LEROUXEL O, HAN K H . ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. The Plant Journal, 2007,50(6):1035-1048.
doi: 10.1111/j.1365-313X.2007.03109.x pmid: 17565617 |
| [10] |
ZHONG R, DEMURA T, YE Z H . SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers ofArabidopsis. The Plant Cell, 2006,18(11):3158-3170.
doi: 10.1105/tpc.106.047399 pmid: 17114348 |
| [11] |
YAMAGUCHI M, KUBO M, FUKUDA H, DEMURA T . Vascular-related NAC-domain7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. The Plant Journal, 2008,55(4):652-664.
doi: 10.1111/j.1365-313X.2008.03533.x pmid: 18445131 |
| [12] | ODA-YAMAMIZO C, MITSUDA N, SAKAMOTO S, OGAWA D, OHME-TAKAGI M, OHMIYA A . ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Scientific Reports, 2016,6:23609. |
| [13] | TAKASAKI H, MARUYAMA K, TAKAHASHI F, FUJITA M, YOSHIDA T, NAKASHIMA K, MYOUGA F, TOYOOKA K, YAMAGUCHI-SHINOZAKI K, SHINOZAKI K . SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. The Plant Journal, 2015,84(6):1114-1123. |
| [14] | ZHU Y, YAN J W, LIU W J, LIU L, SHENG Y, SUN Y, LI Y Y, SCHELLER H, JIANG M Y, HOU X L, NI L, ZHANG A Y . Phosphorylation of a NAC transcription factor by a calcium/ calmodulin-dependent protein kinase regulates abscisic acid-induced antioxidant defense in maize. Plant Physiology, 2016,171(3):1651-1664. |
| [15] |
ZHOU H, WANG K L, WANG H L, GU C, DARE A, ESPLEY R, HE H P, ALLAN A, HAN Y P . Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. The Plant Journal, 2015,82(1):105-121.
doi: 10.1111/tpj.12792 pmid: 25688923 |
| [16] | 安建平, 宋来庆, 赵玲玲, 由春香, 王小非, 郝玉金 . 苹果愈伤组织超表达MdNAC029促进花青苷积累. 园艺学报, 2018,45(5):845-854. |
| AN J P, SONG L Q, ZHAO L L, YOU C X, WANG X F, HAO Y J . Overexpression of MdNAC029 promotes anthocyanin accumulation in apple calli. Acta Horticulturae Sinica, 2018,45(5):845-854. (in Chinese) | |
| [17] |
NURUZZAMAN M, MANIMEKALAI R, SHRRONI A M . Genome-wide analysis of NAC transcription factor family in rice. Gene, 2010,465(1/2):30-44.
doi: 10.1016/j.gene.2010.06.008 pmid: 20600702 |
| [18] |
KIM S G, LEE S, SEO P J, KIM S K, KIM J K, PARK C M . Genome-scale screening and molecular characterization of membrane- bound transcription factors in Arabidopsis and rice. Genomics, 2010,95(1):56-65.
doi: 10.1016/j.ygeno.2009.09.003 pmid: 19766710 |
| [19] |
SONG Y, LIU H D, ZHOU Q, ZHANG H J, ZHANG Z D, LI Y D, WANG H B, LIU F Z . High-throughput sequencing of highbush blueberry transcriptome and analysis of basic helix-loop-helix transcription factors. Journal of Integrative Agriculture, 2017,16(3):591-604.
doi: 10.1016/S2095-3119(16)61461-2 |
| [20] | YIN X R, ALLAN A C, CHEN K S, FERGUSON I B . Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiology, 2010,153(3):1280-1292. |
| [21] |
JEFFERSON R A, KAVANAGH T A, BEVAN M W . GUS fusions:β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal, 1987,6(13), 3901-3907.
doi: 10.1089/dna.1987.6.583 pmid: 3327686 |
| [22] | PERTUZATTI P B, BARCIA M T , REBELLO L P G, GÓMEZ- ALONSO S, DUARTE R M T, DUARTE M C T, GODOY H T, HERMOSÍN-GUTIÉRREZ I .Antimicrobial activity and differentiation of anthocyanin profiles of rabbiteye and highbush blueberry using HPLC-DAD-ESI-MS and multivariate analysis. Journal of Functional Foods, 2016,26:506-516. |
| [23] |
ZHU X Y, CHEN J Y, XIE Z K, GAO J, REN G D, GAO S, ZHOU X, KUAI B K . Jasmonic acid promotes degreening via MYB2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. The Plant Journal, 2015,84(3):597-610.
doi: 10.1111/tpj.13030 pmid: 26407000 |
| [24] |
LI S, GAO J, YAO L Y, REN G D, ZHU X Y, GAO S, QIU K, ZHOU X, KUAI B K . The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark- induced leaf senescence. Plant Cell Reports, 2016,35(8):1729-1741.
doi: 10.1007/s00299-016-1991-1 pmid: 27154758 |
| [25] |
HUANG J C, PIATER L A, DUBERY I A . The NAC transcription factor gene ANAC072 is differentially expressed in Arabidopsis thaliana in response to microbe-associated molecular pattern (MAMP) molecules. Physiological and Molecular Plant Pathology, 2012,80(80):19-27.
doi: 10.1016/j.pmpp.2012.07.002 |
| [26] |
TRAN L S, NAKASHIMA K, SAKUMA Y, SIMPSON S D, FUJITA Y, MARUYAMA K, FUJITA M, SEKI M, SHINOZAKI K, YAMAGUCHI-SHINOZAKI K . Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell, 2004,16(9):2481-2498.
doi: 10.1105/tpc.104.022699 pmid: 15319476 |
| [27] |
LI X Y, SUN H Y, PEI J B, DONG Y Y, WANG F W, CHEN H, SUN Y P, WANG N, LI H Y, LI Y D . De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidant. Gene, 2012,511(1):54-61.
doi: 10.1016/j.gene.2012.09.021 pmid: 22995346 |
| [28] |
SUN H Y, LIU Y S, GAI Y Z, GENG J M, CHEN L, LIU H D, KANG L M, TIAN Y W, LI Y D . De novo sequencing and analysis of the cranberry fruit transcriptome to identify putative genes involved in flavonoid biosynthesis, transport and regulation. BMC Genomics, 2015,16(1):652.
doi: 10.1186/s12864-015-1842-4 pmid: 4556307 |
| [29] |
BOREVITZ J O, XIA Y J, BLOUNT J, DIXON R, LAMB C . Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell, 2000,12(12):2383-2393.
doi: 10.2307/3871236 pmid: 11148285 |
| [30] |
TAKOS A, JAFFÉ F, JACOB S, BOGS J, ROBINSON S, WALKER A . Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiology, 2006,142(3):1216-1232.
doi: 10.1104/pp.106.088104 pmid: 17012405 |
| [31] | YAO G F, MING M L, ALLAN A, GU C, LI L T, WU X, WANG R Z, CHANG Y J, QI K J, ZHANG S L, WU J . Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. The Plant Journal, 2017,92:437-451. |
| [32] | JAAKOLA L, POOLE M O, JONES M ,KÄMÄRÄINEN- KARPPINEN T,KOSKIMÄKI J,HOHTOLA A,HÄGGMAN H D,FRASER P,MANNING K J,KING G. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiology, 2010,153(4):1619-1629. |
| [33] | SAKURABA Y, KIM Y S, HAN S H, LEE B D, PAEK N C . The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. The Plant Cell, 2015,27(6):1771-1787. |
| [34] | QU Y T, DUAN M, ZHANG Z Q, DONG J L, WANG T . Overexpression of the Medicago falcata NAC transcription factor MfNAC3 enhances cold tolerance in Medicago truncatula. Environmental and Experimental Botany, 2016,129:67-76. |
| [1] | 李世佳,吕紫敬,赵锦. 枣R2R3-MYB亚家族基因鉴定及其在果实发育中的表达分析[J]. 中国农业科学, 2022, 55(6): 1199-1212. |
| [2] | 赖春旺, 周小娟, 陈燕, 刘梦雨, 薛晓东, 肖学宸, 林文忠, 赖钟雄, 林玉玲. 龙眼乙烯合成途径基因鉴定及响应ACC处理的分析[J]. 中国农业科学, 2022, 55(3): 558-574. |
| [3] | 陈婷婷, 符卫蒙, 余景, 奉保华, 李光彦, 符冠富, 陶龙兴. 彩色稻叶片光合特征及其与抗氧化酶活性、花青素含量的关系[J]. 中国农业科学, 2022, 55(3): 467-478. |
| [4] | 由玉婉,张雨,孙嘉毅,张蔚. ‘月月粉’月季NAC家族全基因组鉴定及皮刺发育相关成员的筛选[J]. 中国农业科学, 2022, 55(24): 4895-4911. |
| [5] | 孙保娟,汪瑞,孙光闻,王益奎,李涛,宫超,衡周,游倩,李植良. 转录组及代谢组联合解析茄子果色上位遗传效应[J]. 中国农业科学, 2022, 55(20): 3997-4010. |
| [6] | 袁景丽,郑红丽,梁先利,梅俊,余东亮,孙玉强,柯丽萍. 花青素代谢对陆地棉叶片和纤维色泽呈现的影响[J]. 中国农业科学, 2021, 54(9): 1846-1855. |
| [7] | 刘闯,高振,姚玉新,杜远鹏. 葡萄钾离子转运基因VviHKT1;7在盐胁迫下的功能鉴定[J]. 中国农业科学, 2021, 54(9): 1952-1963. |
| [8] | 吕士凯, 马小龙, 张敏, 邓平川, 陈春环, 张宏, 刘新伦, 吉万全. 小麦TaNAC基因基于可变剪切和microRNA的转录后调控分析[J]. 中国农业科学, 2021, 54(22): 4709-4727. |
| [9] | 刘有春,刘威生,王兴东,孙斌,刘修丽,杨艳敏,魏鑫,杨玉春,张舵,刘成,李天忠. 基于简化基因组测序的越橘杂交后代鉴定[J]. 中国农业科学, 2021, 54(2): 370-378. |
| [10] | 黄金凤,吕天星,王寻,王颖达,王冬梅,闫忠业,刘志. 苹果LRR-RLK基因家族鉴定和表达分析[J]. 中国农业科学, 2021, 54(14): 3097-3112. |
| [11] | 崔虎亮,贺霞,张前. 不同牡丹品种开花期间花瓣花青素和类黄酮组成的动态变化[J]. 中国农业科学, 2021, 54(13): 2858-2869. |
| [12] | 马建, 李丛丛, 黄亚婷, 谢雨黎, 程玲玲, 王建设. 甜瓜种皮颜色控制基因CmSC1的精细定位及候选基因分析[J]. 中国农业科学, 2021, 54(10): 2167-2178. |
| [13] | 郝小燕,牟春堂,乔栋,张暄梓,杨文军,赵俊星,张春香,张建新. 葡萄籽原花青素对羔羊瘤胃发酵、血清炎症及抗氧化指标的影响[J]. 中国农业科学, 2021, 54(10): 2239-2248. |
| [14] | 邵晨冰,黄志楠,白雪滢,王云鹏,段伟科. 辣椒HD-Zip基因家族鉴定、系统进化及表达分析[J]. 中国农业科学, 2020, 53(5): 1004-1017. |
| [15] | 许明,林世强,倪冬昕,伊恒杰,刘江洪,杨志坚,郑金贵. 藤茶查尔酮合成酶基因AgCHS1的克隆及功能鉴定[J]. 中国农业科学, 2020, 53(24): 5091-5103. |
|
||