













中国农业科学 ›› 2022, Vol. 55 ›› Issue (22): 4356-4372.doi: 10.3864/j.issn.0578-1752.2022.22.003
王娟1( ),陈皓宁1,2,石大川3,于天一1,闫彩霞1,孙全喜1,苑翠玲1,赵小波1,牟艺菲1,王奇1,李春娟1(
),陈皓宁1,2,石大川3,于天一1,闫彩霞1,孙全喜1,苑翠玲1,赵小波1,牟艺菲1,王奇1,李春娟1( ),单世华1(
),单世华1( )
)
收稿日期:2021-07-12
接受日期:2022-09-08
出版日期:2022-11-16
发布日期:2022-12-14
通讯作者:
李春娟,单世华
作者简介:王娟,E-mail:基金资助:
WANG Juan1( ),CHEN HaoNing1,2,SHI DaChuan3,YU TianYi1,YAN CaiXia1,SUN QuanXi1,YUAN CuiLing1,ZHAO XiaoBo1,MOU YiFei1,WANG Qi1,LI ChunJuan1(
),CHEN HaoNing1,2,SHI DaChuan3,YU TianYi1,YAN CaiXia1,SUN QuanXi1,YUAN CuiLing1,ZHAO XiaoBo1,MOU YiFei1,WANG Qi1,LI ChunJuan1( ),SHAN ShiHua1(
),SHAN ShiHua1( )
)
Received:2021-07-12
Accepted:2022-09-08
Online:2022-11-16
Published:2022-12-14
Contact:
ChunJuan LI,ShiHua SHAN
摘要:
【目的】 氮素在作物生产中对生物量和产量起关键作用。高亲和硝酸盐转运蛋白基因NRT2在植物响应低氮胁迫时被激活并具有维持氮吸收和转运的作用。通过筛选花生低氮耐受相关的NRT2基因并解析其生物学功能,为培育高氮素利用率的花生新品种提供理论参考,最终有助于实现节氮、高效的绿色生产目标。【方法】 检测正常氮浓度及1/20正常氮浓度(15 mmol·L-1)条件下5个具有典型跨膜结构的花生NRT2基因(AhNRT2.4、AhNRT2.5b、AhNRT2.5c、AhNRT2.7a和AhNRT2.7b)的时空表达情况。以花育6309的cDNA为模板,对AhNRT2.7a进行克隆和生物信息学分析,并通过亚细胞定位确定AhNRT2.7a的表达部位。进一步构建异源过表达AhNRT2.7a的拟南芥株系,分别在正常以及低氮胁迫条件下测定其叶绿素含量、氮积累量以及谷氨酰胺合成酶(GS)、谷氨酸合成酶(GOGAT)、硝酸还原酶(NR)、亚硝酸还原酶(NiR)、谷氨酸脱氢酶(GDH)5个氮代谢关键酶的活性。【结果】 5个NRT2基因中有4个NRT2基因在花生响应低氮胁迫条件下大量表达。其中,AhNRT2.7a能够响应低氮胁迫,并在花生茎和叶中高表达。获得AhNRT2.7a的cDNA序列,全长为1 380 bp,编码459个氨基酸。蛋白结构分析显示其为具有12个典型跨膜结构域的膜蛋白。该氨基酸序列与栽培种花生(Arachis hypogaea L.)相似性高达99.56%,其次是野生亲本AA和BB基因组花生。亚细胞定位显示其主要定位于细胞质膜上。构建异源过表达AhNRT2.7a的转基因拟南芥植株,在不同供氮条件下,成熟叶和幼叶叶片的叶绿素相对含量均显著高于野生型拟南芥。同时,对上述5个氮代谢相关酶活性以及氮、磷、钾积累量测定显示,转基因植株中2个氮代谢相关酶(GS和NR)的酶活性以及氮积累量均比野生型拟南芥有显著升高。【结论】 花生中4个NRT2基因响应低氮胁迫,其中AhNRT2.7a能够提高植物氮代谢过程的氮素利用率。AhNRT2.7a的表达在促进氮代谢过程的同时,促使碳代谢作用的进一步加强。因此,AhNRT2.7a适合作为花生氮素高效利用为目的的候选基因。
王娟,陈皓宁,石大川,于天一,闫彩霞,孙全喜,苑翠玲,赵小波,牟艺菲,王奇,李春娟,单世华. 花生高亲和硝酸盐转运蛋白基因AhNRT2.7a响应低氮胁迫的功能研究[J]. 中国农业科学, 2022, 55(22): 4356-4372.
WANG Juan,CHEN HaoNing,SHI DaChuan,YU TianYi,YAN CaiXia,SUN QuanXi,YUAN CuiLing,ZHAO XiaoBo,MOU YiFei,WANG Qi,LI ChunJuan,SHAN ShiHua. Functional Analysis of AhNRT2.7a in Response to Low-Nitrogen in Peanut[J]. Scientia Agricultura Sinica, 2022, 55(22): 4356-4372.
表1
5个NRT2基因及其引物序列"
| 基因名称 Gene name | 正向引物序列 F-primer (5′-3′) | 反向引物序列 R-primer (5′-3′) |
| AhNRT2.4 | TGCCCTTTTGTATGCAACATTT | CTCATACCAAACAACCGGGC |
| AhNRT2.5b | TCTGTGTTTGTTCAAGCTGC | CCTCCTCCTGTCATTCCTGA |
| AhNRT2.5c | CTCTGTGTTTGTTCAAGCTG | TCCTCCTGTCATTCCTGATA |
| AhNRT2.7a | ACGGTCAAGATCTCCCTTCT | CACGCACCACAACAACAAAT |
| AhNRT2.7b | GGTTCTGGGACTGCTGTATG | ACCCCGAACCTGTCATAGAA |
| Actin | TTGGAATGGGTCAGAAGGATGC | AGTGGTGCCTCAGTAAGAAGC |
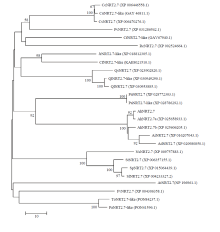
图4
AhNRT2.7a与其他物种同源蛋白的系统进化树分析 CcNRT2.7:中华辣椒,XP_006446558.1;CsNRT2.7:黄瓜,XP_006470276.1;PvNRT2.7:开心果,XP_031286542.1;RcNRT2.7:月季,XP_002524664.1;JrNRT2.7:核桃,XP_002524664.1;QsNRT2.7:栓皮栎,XP_023902820.1;QlNRT2.7:白栎,XP_030953885.1;PaNRT2.7:白牧豆树,XP_028772303.1;AhNRT2.7a:栽培种花生,XP_025658933.1,XP_025606205.1;AiNRT2.7a:野生花生,XP_016207043.1;AdNRT2.7a:野生花生,XP_020980850.1;AtNRT2.7:拟南芥,NP_196961.1;NsNRT2.7:美花烟草,XP_009757883.1;StNRT2.7:马铃薯,XP_006357155.1;SpNRT2.7:潘那利番茄,XP_015064439.1;SlNRT2.7:番茄,XP_004233327.2;FvNRT2.7:野草莓,XP_004306358.1;PaNRT2.7-like:白牧豆,PON41596.1"
| [1] | 廖伯寿. 我国花生生产发展现状与潜力分析. 中国油料作物学报, 2020, 42(2): 161-166. |
| LIAO B S. Analysis on development status and potential of peanut production in China. Chinese Journal of Oil Crops Sciences, 2020, 42(2): 161-166. (in Chinese) | |
| [2] | 李向东, 万勇善, 张高英, 吴爱荣, 马晓东. 夏花生覆膜对根瘤中固氮酶和叶片硝酸还原酶活性影响的研究. 作物学报, 1996, 22(1): 96-100. |
| LI X D, WAN Y S, ZHANG G Y, WU A R, MA X D. Effects of film mulching on nitrogenase and nitrate reductase activities in root nodules of peanut in summer. Acta Agronomica Sinica, 1996, 22(1): 96-100. (in Chinese) | |
| [3] | 李向东, 王晓云, 张高英, 万勇善, 李军. 花生衰老的氮素调控. 中国农业科学, 2000, 33(5): 1-7. |
| LI X D, WANG X Y, ZHANG G Y, WAN Y S, LI J. Regulation of nitrogen in peanut senescence. Scientia Agricultura Sinica, 2000, 33(5): 1-7. (in Chinese) | |
| [4] |
ROY S, LIU W, NANDETY R S, CROOK A, MYSORE K S, LISLARIU C I, FRUGOLI J, DICKSTEIN R, UDVARDI M K. Celebrating 20 years of genetic discoveries in Legume nodulation and symbiotic nitrogen fixation. The Plant Cell, 2019, 32(1): 15-41.
doi: 10.1105/tpc.19.00279 |
| [5] |
WANG Y Y, CHENG Y H, CHEN K E, TSAY Y F. Nitrate transport, signaling, and use efficiency. Annual Review of Plant Biology, 2018, 69: 85-122.
doi: 10.1146/annurev-arplant-042817-040056 |
| [6] | 刘颖, 张佳蕾, 李新国, 张正, 万书波. 豆科作物氮素高效利用机制研究进展. 中国油料作物学报, 2022, 44(3): 476-482. |
| LIU Y, ZHANG J L, LI X G, ZHANG Z, WAN S B. Research progress on nitrogen efficient utilization mechanism of leguminous crops. Chinese Journal of Oil Crop Sciences, 2022, 44(3): 476-482. (in Chinese) | |
| [7] |
CHEN C Z, LU X F, LI J Y, YI H Y, GONG J M. Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiology, 2012, 159: 1582-1590.
doi: 10.1104/pp.112.199257 |
| [8] | 于飞, 施卫明. 近10年中国大陆主要粮食作物氮肥利用率分析. 土壤学报, 2015, 52(6): 1311-1324. |
| YU F, SHI W M. Analysis of nitrogen use efficiency of main grain crops in mainland China in recent 10 years. Acta Pedologica Sinica, 2015, 52(6): 1311-1324. (in Chinese) | |
| [9] |
WEST P C, GERBER J S, ENGSTROM P M, ENGSTROM P M, MUELLER N D, BRAUMAN K A, CARLSON K M, CASSIDY E S, JOHNSTON M, MACDONALD G K, RAY D K. Leverage points for improving global food security and the environment. Science, 2014, 345(6194): 325-328.
doi: 10.1126/science.1246067 pmid: 25035492 |
| [10] | 张卫峰, 马林, 黄高强, 武良, 陈新平, 张福锁. 中国氮肥发展、贡献和挑战. 中国农业科学, 2013, 46(15): 3161-3171. |
| ZHANG W F, MA L, HUANG G Q, WU L, CHEN X P, ZHANG F S. Nitrogen fertilizer development, contribution and challenge in China. Scientia Agricultura Sinica, 2013, 46(15): 3161-3171. (in Chinese) | |
| [11] |
LERAN S, VARALA K, BOYER J C, CHIURAZZI M, CRAWFORD N, DANIEL-VEDELE F, DAVID L, DICKSTEIN R, FERNANDEZ E. A unified nomenclature of NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science, 2014, 19: 5-9.
doi: 10.1016/j.tplants.2013.08.008 |
| [12] |
GALVAN A, FERNANDEZ E. Eukaryotic nitrate and nitrite transporter. Cellular and Molecular Life Science, 2001, 58: 225-233.
doi: 10.1007/PL00000850 |
| [13] | 张合琼, 张汉马, 梁永书, 南文斌. 植物硝酸盐转运蛋白研究进展. 植物生理学报, 2016, 52(336): 4-12. |
| ZHANG H Q, ZHANG H M, LIANG Y S, NAN W B. Research progress of nitrate transporter in plants. Plant Physiology Journal, 2016, 52(336): 4-12. (in Chinese) | |
| [14] |
OKAMOT M, KUMAR A, LI W B, WANG Y, SIDDIQI M Y, CRAWFORD N M, GLASS A D M. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiology, 2006, 140: 1036-1046.
doi: 10.1104/pp.105.074385 |
| [15] |
FORDE B G. Nitrate transporters in plants: Structure, function and regulation. Biochimica et Biophysica Acta-Biomembranes, 2000, 1465(1/2): 219-235.
doi: 10.1016/S0005-2736(00)00140-1 |
| [16] |
ORSEL M, CHOPIN F, LELEU O, SMITH S H, KRAPP A, DANIEL-VEDELE F, MILLER A J. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiology, 2006, 142: 1304-1317.
doi: 10.1104/pp.106.085209 |
| [17] |
LEZHNEVA L, KIBA T, FERIA-BOURRELLIER A B, LAFOUGE F, BUOTET-MERCEY S, ZOUFAN P, SAKAKIBARA H, DANIEL-VEDELE F, KRAPP A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant Journal for Cell and Molecular Biology, 2014, 80(2): 230-241.
doi: 10.1111/tpj.12626 |
| [18] | PENUELAS J, SARDANS J. The global nitrogen-phosphorus imbalance. Science, 2022, 375: 6578. |
| [19] |
LIU J, CHEN F, OLOKHNUUD C, GLASS A D M, TONG Y, ZHANG F, MI G. Root size and nitrogen uptake activity in two maize (Zea mays) inbred lines differing in nitrogen use efficiency. Journal of Plant Nutrition and Soil Science, 2009, 172(2): 230-236.
doi: 10.1002/jpln.200800028 |
| [20] |
TANG Z, FAN X, LI Q, FENG H, MILLER A J, SHEN Q, XU G. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiology, 2012, 160(4): 2052-2063.
doi: 10.1104/pp.112.204461 pmid: 23093362 |
| [21] | FAN X, TANG Z, TAN Y, ZHANG Y, LUO B, YANG M, XU, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proceedings of the National Academy of Sciences of the USA, 2016, 113(26): 7118-7123. |
| [22] |
PELLIZZARO A, CLOCHARD T, PLANCHT E, LIMAMI A M, MORERLEORER-LE PAVEN M C. Identification and molecular characterization of Medicago truncatula NRT2 and NAR2 families. Physiologia Plantarum, 2015, 154(2): 256-269.
doi: 10.1111/ppl.12314 |
| [23] |
VON WITTGENSTEIN N J, LE C H, HAWKINS B J, EHLTING J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evolutionary Biology, 2014, 14: 11.
doi: 10.1186/1471-2148-14-11 pmid: 24438197 |
| [24] | 宋田丽, 周建建, 徐晨曦, 蔡晓峰, 戴绍军, 王全华, 王小丽. 植物硝酸盐转运蛋白功能及表达调控研究进展. 上海师范大学学报(自然科学版), 2017, 46(5): 740-750. |
| SONG T L, ZHOU J J, XU C X, CAI X F, DAI S J, WANG Q H, WANG X L. Research progress on function and expression regulation of Nitrate transporter in plants. Journal of Shanghai Normal University (Nature Sciences), 2017, 46(5): 740-750. (in Chinese) | |
| [25] |
CHEN J, ZHANG Y, TAN Y, ZHANG M, ZHU L, XU G, FAN X. Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT 2.1 expression with the OsNAR 2.1 promoter. Plant Biotechnology Journal, 2016, 14(8): 1705-1715.
doi: 10.1111/pbi.12531 |
| [26] |
ZHUANG W J, CHEN H, YANG M, WANG J P, PANDEY M K, ZHANG C, CHANG W C, ZHANG L S, ZHANG X T. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nature Genetics, 2019, 51(5): 865-876.
doi: 10.1038/s41588-019-0402-2 pmid: 31043757 |
| [27] |
BERTIOLI D J, JENKINS J, CLEVENGER J, DUDCHENKO O, GAO D Y, SEIJO G, LEAL-BERTIOLI S C M, REN L H, FARMER A D, PANDEY M K, SAMOLUK S S, ABERNATHY B, AGARWA G, BALLÉN-TABORDA C, CAMERON C, CAMPBELL J, CHAVARRO C, CHITIKINENI A, CHU Y, DASH S, BAIDOURI M E, GUO B Z, HUANG W, KIM K D, KORANI W, LANCIANO S, LUI C G, MIROUZE M, MORETZSOHN M C, PHAM M, SHIN J H, SHIRASAWA K, SINHAROY S, SREEDASYAM A, WEEKS N T, ZHANG X Y, ZHENG Z, SUN Z Q, FROENICKE L, AIDEN E L, MICHELMORE R I, VARSHNEY R K, HOLBROOK C C, CANNON E K S, SCHEFFLER B E, GRIMWOOD J, OZIAS-AKINS P, CANNON S B, JACKSON S A, SCHMUTZ J. The genome sequence of segmental allotetraploid peanut Arachis hypogaea L. Nature Genetics, 2019, 51(5): 877-884.
doi: 10.1038/s41588-019-0405-z |
| [28] | CLEVENGERl J, CHU Y, SCHEFFLER B, OZIAS-AKINS P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Frontiers in Plant Science, 2016, 7: 1446. |
| [29] |
ZHAO Y H, MA J J, LI M, DENG L, LI G H, XIA H, ZHAO S Z, HOU L, LI P C, MA C L, YUAN M, REN L, GU J Z, GUO B Z, ZHAO C Z, WANG X J. Whole-genome resequencing-based QTL-seq identified AhTc1 gene encoding a R2R3-MYB transcription factor controlling peanut purple testa colour. Plant Biotechnology Journal, 2020, 18: 96-105.
doi: 10.1111/pbi.13175 |
| [30] | 王娟, 石大川, 陈皓宁, 吴丽青, 闫彩霞, 陈静, 赵小波, 孙全喜, 苑翠玲, 牟艺菲, 单世华, 李春娟. 花生高亲和硝酸盐转运蛋白基因家族生物信息学分析. 中国油料作物学报, 2022, 44(2): 316-323. |
| WANG J, SHI D C, CHEN H N, WU L Q, YAN C X, CHEN J, ZHAO X B, SUN Q X, YUAN C L, MU Y F, SHAN S H, LI C J. Bioinformatics analysis of high affinity nitrate transporter gene family in Peanut. Chinese Journal of Oil Crop Sciences, 2022, 44(2): 316-323. (in Chinese) | |
| [31] |
LERAN S, VARALA K, BOYER J C, CHIURAZZI M, LACOMBE B. A unified nomenclature of NITRATE TRANSPORTER1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science, 2014, 19: 5-9.
doi: 10.1016/j.tplants.2013.08.008 |
| [32] |
WANG J, YAN C X, LI Y, LI C J, ZHAO X B, YUAN C L, SUN Q X, SHAN S H. GWAS discovery of candidate genes for yield-related traits in peanut and support from earlier QTL mapping studies. Genes, 2019, 10(10): 803.
doi: 10.3390/genes10100803 |
| [33] |
TANG W J, YE J, YAO X M, ZHAO P Z, XUAN W, TIAN T L, ZHANG Y Y, XU S, AN H Z, CHEN G M. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nature Communications, 2019, 10(1): 5279.
doi: 10.1038/s41467-019-13187-1 pmid: 31754193 |
| [34] | 武姣娜, 魏晓东, 李霞, 张金飞, 谢寅峰. 植物氮素利用率的研究进展. 植物生理学报, 2018, 54(9): 1401-1408. |
| WU J N, WEI X D, LI X, ZHANG J F, XIE Y F. Research progress of nitrogen use efficiency in plants. Plant Physiology Journal, 2018, 54(9): 1401-1408. (in Chinese) | |
| [35] |
TENG W, HE X, TONG Y. Transgenic approaches for improving use efficiency of nitrogen, phosphorus and potassium in crops. Journal of Integrative Agriculture, 2017, 16(12): 2657-2673.
doi: 10.1016/S2095-3119(17)61709-X |
| [36] |
LI H, HU B, CHU C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. Journal of Experimental Botany, 2017, 68(10): 2477-2488.
doi: 10.1093/jxb/erx101 |
| [37] |
GABRIEL K, NIGEL M C, GLORIA M C, TSAY Y F. Nitrate signaling: Adaptation to fluctuating environments. Current Opinion in Plant Biology, 2015, 13(3): 265-272.
doi: 10.1016/j.pbi.2009.12.003 |
| [38] |
KECHID M, DESBROSSES G, ROKHSI W, VAROQUAN F, DJEKOUN A, TOURAINE B. The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth- promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM 196. New Phytologist, 2013, 198(2): 514-524.
doi: 10.1111/nph.12158 |
| [39] | 轩红梅, 王永华, 魏利婷, 杨莹莹, 王利娜, 康国章, 郭天财. 小麦幼苗叶片中硝酸盐转运蛋白NRT1和NRT2家族基因对氮饥饿响应的表达分析. 麦类作物学报, 2014, 34(8): 1019-1028. |
| XUAN H M, WANG Y H, WEI L T, YANG Y Y, WANG L N, KANG G Z, GUO T C. Expression analysis of nitrate transporter NRT1and NRT2 family genes in response to nitrogen starvation in wheat seedling leaves. Journal of Triticeae Crops, 2014, 34(8): 1019-1028. (in Chinese) | |
| [40] |
WEI J, ZHANG Y, FENG H, QU H, FAN X, YAMAJI N, XU G. OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. Journal of Experimental Botany, 2018, 69(5): 1095-1107.
doi: 10.1093/jxb/erx486 |
| [41] | LIU Y Y, SHAO L B, ZHOU J, LI R C, PANDEY M K, HAN Y, CUI F, ZHANG J L, GUO F, CHEN J, SHAN S H, FAN G Y, ZHANG H, SEIM I, LIU X, LI X G, VARSHNEY R K, LI G W, WAN S B. Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. Journal of Advanced Research, 2022, https://doi.org/10.1016/j.jare.2022.01.016 |
| [42] | 郑永美, 周丽梅, 郑亚萍, 吴正锋, 孙学武, 于天一, 沈浦, 王才斌. 花生主要碳代谢指标与根瘤固氮能力的关系. 植物营养与肥料学报, 2021, 27(1): 75-86. |
| ZHENG Y M, ZHOU L M, ZHENG Y P, WU Z F, SUN X W, YU T Y, SHEN P, WANG C B. Relationship between carbon metabolism and nitrogen fixation ability of peanut nodule. Journal of Plant Nutrition and Fertilizer, 2021, 27(1): 75-86. (in Chinese) | |
| [43] | CAMPBELL W H. Nitrate reductase structure, function and regulation: Bridging the gap between biochemistry and physiology. Annual Review of Plant Biology, 1999, 50(1): 277-303. |
| [1] | 吴月,隋新华,戴良香,郑永美,张智猛,田云云,于天一,孙学武,孙棋棋,马登超,吴正锋. 慢生根瘤菌及其与花生共生机制研究进展[J]. 中国农业科学, 2022, 55(8): 1518-1528. |
| [2] | 卞能飞, 孙东雷, 巩佳莉, 王幸, 邢兴华, 金夏红, 王晓军. 花生烘烤食用品质评价及指标筛选[J]. 中国农业科学, 2022, 55(4): 641-652. |
| [3] | 郭灿,岳晓凤,白艺珍,张良晓,张奇,李培武. 花生黄曲霉毒素平衡取样-随机森林风险预警模型的应用研究[J]. 中国农业科学, 2022, 55(17): 3426-3436. |
| [4] | 史晓龙,郭佩,任婧瑶,张鹤,董奇琦,赵新华,周宇飞,张正,万书波,于海秋. 基于花生//高粱间作模式的花生盐胁迫耐受性效应研究[J]. 中国农业科学, 2022, 55(15): 2927-2937. |
| [5] | 郝静,李秀坤,崔顺立,邓洪涛,侯名语,刘盈茹,杨鑫雷,穆国俊,刘立峰. 花生每荚种子数相关性状QTL的定位[J]. 中国农业科学, 2022, 55(13): 2500-2508. |
| [6] | 冯晨,黄波,冯良山,郑家明,白伟,杜桂娟,向午燕,蔡倩,张哲,孙占祥. 不同配置对辽西玉米‖花生间作系统氮素吸收利用的影响[J]. 中国农业科学, 2022, 55(1): 61-73. |
| [7] | 孟鑫浩,邓洪涛,李丽,崔顺立,Charles Y.Chen,侯名语,杨鑫雷,刘立峰. 栽培种花生株型相关性状的QTL定位[J]. 中国农业科学, 2021, 54(8): 1599-1612. |
| [8] | 陈歌,曹立冬,许春丽,赵鹏跃,曹冲,李凤敏,黄啟良. 溶剂蒸发法制备丙硫菌唑微囊及其性能研究[J]. 中国农业科学, 2021, 54(4): 754-767. |
| [9] | 荆丹, 岳晓凤, 白艺珍, 郭灿, 丁小霞, 李培武, 张奇. 花生黄曲霉侵染力[J]. 中国农业科学, 2021, 54(23): 5008-5020. |
| [10] | 顾博文,杨劲峰,鲁晓玲,吴怡慧,李娜,刘宁,安宁,韩晓日. 连续施用生物炭对花生不同生育时期叶绿素荧光特性的影响[J]. 中国农业科学, 2021, 54(21): 4552-4561. |
| [11] | 薛华龙,娄梦玉,李雪,王飞,郭彬彬,郭大勇,李海港,焦念元. 施磷水平对不同茬口下冬小麦生长发育及产量的影响[J]. 中国农业科学, 2021, 54(17): 3712-3725. |
| [12] | 张毛宁,黄冰艳,苗利娟,徐静,石磊,张忠信,孙子淇,刘华,齐飞艳,董文召,郑峥,张新友. 巢式杂交分离群体的花生籽仁性状的主基因+多基因混合遗传模型分析[J]. 中国农业科学, 2021, 54(13): 2916-2930. |
| [13] | 胡廷会,成良强,王军,吕建伟,饶庆琳. 不同基因型花生耐荫性评价及其鉴定指标的筛选[J]. 中国农业科学, 2020, 53(6): 1140-1153. |
| [14] | 徐志军,赵胜,徐磊,胡小文,安东升,刘洋. 基于RNA-seq数据的栽培种花生SSR位点鉴定和标记开发[J]. 中国农业科学, 2020, 53(4): 695-706. |
| [15] | 郑福丽,刘苹,李国生,张柏松,李燕,魏建林,谭德水. 有机-无机肥协同调控小麦-玉米两熟作物产量及土壤培肥效应[J]. 中国农业科学, 2020, 53(21): 4355-4364. |
|
||