













中国农业科学 ›› 2023, Vol. 56 ›› Issue (2): 264-274.doi: 10.3864/j.issn.0578-1752.2023.02.005
刘瑞1( ),赵羽涵1,付忠举1,顾欣怡1,王艳霞1,靳学慧1,杨莹1,吴伟怀2,张亚玲1(
),赵羽涵1,付忠举1,顾欣怡1,王艳霞1,靳学慧1,杨莹1,吴伟怀2,张亚玲1( )
)
收稿日期:2022-08-02
接受日期:2022-09-06
出版日期:2023-01-16
发布日期:2023-02-07
通讯作者:
张亚玲,E-mail:作者简介:刘瑞,E-mail:基金资助:
LIU RUI1( ),ZHAO YuHan1,FU ZhongJu1,GU XinYi1,WANG YanXia1,JIN XueHui1,YANG Ying1,WU WeiHuai2,ZHANG YaLing1(
),ZHAO YuHan1,FU ZhongJu1,GU XinYi1,WANG YanXia1,JIN XueHui1,YANG Ying1,WU WeiHuai2,ZHANG YaLing1( )
)
Received:2022-08-02
Accepted:2022-09-06
Online:2023-01-16
Published:2023-02-07
摘要:
【目的】了解不同稻瘟病菌(Magnaporthe oryzae)菌株群体中PWL基因家族的分布和变异特征,为研究不同稻瘟病菌群体的遗传多样性及特异性提供依据。【方法】通过参考NCBI中公布的无毒基因序列分别对PWL基因家族的启动子区和CDS区设计特异性引物共8对,对2020年采自黑龙江省和海南省不同地区的397个稻瘟病菌单孢分离菌株提取DNA,进行无毒基因的PCR扩增并通过琼脂糖凝胶电泳检测,从检测结果中分别选取囊括不同地区的代表菌株对扩增片段进行测序分析。测序结果与NCBI中相应无毒基因启动子区和CDS区的碱基与氨基酸序列进行比较分析。【结果】在PCR电泳检测结果中,PWL1在所有菌株中均未检测出;PWL2、PWL3和PWL4在黑龙江省和海南省中均扩增出特异性片段,说明这3种基因在两省中均有分布并分别存在不同的分布频率与变异类型。其中PWL2在黑龙江省和海南省中分布频率最高,分别为98.14%和100%;PWL3和PWL4在两省中的分布频率具有显著差异,这2种基因在黑龙江菌株中频率分别为89.30%和82.79%,而在海南菌株中均为5.49%。通过对无毒基因组合进行分析,结果显示,组合类型可分为6种,分别为PWL(f)、PWL2、PWL3、PWL2+PWL3、PWL2+PWL4、PWL2+PWL3+PWL4。其中黑龙江菌株含有所有组合类型,海南菌株仅含有2种,说明黑龙江菌株与海南菌株相比无毒基因型种类较为丰富。通过对PWL基因家族的PCR产物测序发现,PWL基因家族在启动子区和CDS区变异位点丰富,以点突变和缺失为主要变异类型划分为9种,且不同群体来源菌株的变异类型具有特异性和一致性。其中PWL2检测出5种变异类型PWL2-(1,2,3,4,5),碱基序列变化导致氨基酸序列发生错义突变;PWL3和PWL4分别检测出2种变异类型,分别为PWL3-(1,2)和PWL4-(1,2),均发生移码突变使后面氨基酸发生变化。【结论】不同群体来源的稻瘟病菌菌株中PWL基因家族的分布和变异类型具有地域差异性,且变异位点丰富。
刘瑞, 赵羽涵, 付忠举, 顾欣怡, 王艳霞, 靳学慧, 杨莹, 吴伟怀, 张亚玲. 黑龙江省和海南省PWL基因家族在稻瘟病菌中的分布及变异[J]. 中国农业科学, 2023, 56(2): 264-274.
LIU RUI, ZHAO YuHan, FU ZhongJu, GU XinYi, WANG YanXia, JIN XueHui, YANG Ying, WU WeiHuai, ZHANG YaLing. Distribution and Variation of PWL Gene Family in Rice Magnaporthe oryzae from Heilongjiang Province and Hainan Province[J]. Scientia Agricultura Sinica, 2023, 56(2): 264-274.
表1
黑龙江省和海南省稻瘟病菌菌株"
| 省份Province | 采集地点Sampling location | 菌株编号<BOLD>S</BOLD>train number | 菌株数Number of strains |
|---|---|---|---|
| 黑龙江省(215a) Heilongjiang Province(215a) | 绥化北林区Beilin District, Suihua | BL20001-BL20006 | 6 |
| 绥化庆安Qing’an, Suihua | QA20007-QA20009 | 3 | |
| 绥化青冈Qinggang, Suihua | QG20010-QG20012 | 3 | |
| 伊春铁力Tieli, Yichun | TL20013-TL20018 | 6 | |
| 哈尔滨方正Fangzheng, Harbin | FZ20019-FZ20044 | 26 | |
| 哈尔滨通河Tonghe, Harbin | TH20045-TH20069 | 25 | |
| 哈尔滨木兰Mulan, Harbin | ML20070-ML20081 | 12 | |
| 哈尔滨依兰Yilan, Harbin | YL20082-YL20085 | 4 | |
| 鹤岗绥滨Suibin, Hegang | SB20086-SB20102 | 17 | |
| 佳木斯富锦Fujin, Jiamusi | FJ20103-FJ20106 | 4 | |
| 佳木斯东风区Dongfeng District, Jiamusi | DF20199-DF20215 | 17 | |
| 鸡西虎林Hulin, Jixi | HL20107-HL20110 | 4 | |
| 鸡西密山Mishan, Jixi | MS20111-MS20169 | 59 | |
| 牡丹江东宁Dongning, Mudanjiang | DN20170-DN20198 | 29 | |
| 海南省(182b) Hainan Province(182b) | 东方感城Gancheng, Dongfang | DF20001-DF20004 | 4 |
| 陵水Lingshui | LS20085-LS20094 | 10 | |
| 文昌铺前Puqian, Wenchang | WC20005-WC20038 | 34 | |
| 定安定城Dingcheng, Dingan | DC20039-DC20065 | 27 | |
| 定安雷鸣Leiming, Dingan | LM20066-LM20084 | 19 | |
| 澄迈金江Jinjiang, Chengmai | CM20095-CM20182 | 88 |
表2
用于扩增无毒基因的引物"
| 无毒基因 Avr-gene | 引物序列 Primer sequence (5°-3°) | 片段大小 Length of targeted fragments (bp) | 登录号GenBank accession number |
|---|---|---|---|
| PWL1 | F1:TTTCACGCCCATTAACGG | 1262 | AB480169.1 |
| R1:ACGCACAACACGAGGT | |||
| F2:GTCGCAAATGGACTAACA | 395 | ||
| R2:AAACCCTATTCCAGCAGT | |||
| PWL2 | F1:TTCGGGCACTCCGTTACT | 521 | U26313.1 |
| R1:CCTCTCTTTCGCCTTTG | |||
| F2:ATGAAATGCAACAACATC | 495 | ||
| R2:CCTCACACTTAAGTTAACAC | |||
| PWL3 | F1:TGCGAGTAAAAGCCTGAA | 548 | U36995.1 |
| R1:GTGGGTGTTGTAATAGCGAT | |||
| F2:GCTGGGAAGCCTGTCACCT | 1412 | ||
| R2:GTTCACGCAACTGGAGGG | |||
| PWL4 | F1:TCCTCTGGAACATGACAATAGT | 385 | U36996.1 |
| R1:TCCGTGGGTGTTGTAGTAGC | |||
| F2:CGGCTCATTGTCTGTCGTAAAGG | 292 | ||
| R2:TTCGCTGGTCTTTACAAACTCCC |

图1
PWL基因家族的PCR扩增电泳图 A:PWL2启动子区的部分扩增结果Partial amplification results of PWL2 promoter region;B:PWL2 CDS区的部分扩增结果Partial amplification of PWL2 CDS region;C:PWL3启动子区的部分扩增结果Partial amplification results of PWL3 promoter region;D:PWL3 CDS区的部分扩增结果Partial amplification of PWL3 CDS region;E:PWL4启动子区的部分扩增结果Partial amplification results of PWL4 promoter region;F:PWL4 CDS区的部分扩增结果Partial amplification of PWL4 CDS region;M:DNA marker DL2000;1:Guy11阳性对照Guy11 positive control;2:阴性对照(ddH2O)Negative control (ddH2O);+:存在Existence;-:缺失Missing"
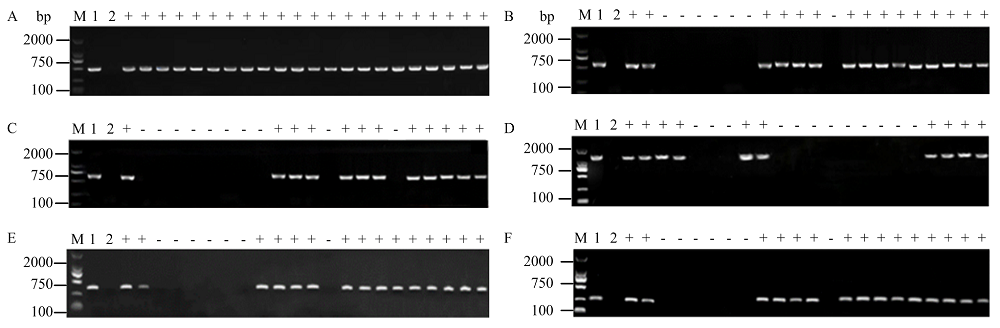
表4
部分菌株PWL2碱基序列比对结果"
| 等位基因型 Allele genotype | 碱基突变位点Base mutation site (bp) | |||||||
|---|---|---|---|---|---|---|---|---|
| -307 | -223 | -169 | -152 | -129 | -73 | -51 | 268 | |
| PWL2 | CGC | TAT | GGT | CAT | TGG | TCG | GAA | GAT |
| PWL2-1 | --T | --- | --- | --- | --- | --- | --- | --- |
| PWL2-2 | --- | --- | --- | --- | --- | --- | --- | A-- |
| PWL2-3 | --- | --- | --- | --- | --- | --- | A-- | A-- |
| PWL2-4 | --- | --G | --G | -T- | G-- | --T | A-- | A-- |
| [1] | 张亚玲, 高清, 赵羽涵, 刘瑞, 付忠举, 李雪, 孙宇佳, 靳学慧. 黑龙江省水稻种质稻瘟病抗性评价及抗瘟基因结构分析. 中国农业科学, 2022, 55(4): 625-640. |
| ZHANG Y L, GAO Q, ZHAO Y H, LIU R, FU Z J, LI X, SUN Y J, JIN X H. Evaluation of rice blast resistance and genetic structure analysis of rice germplasm in Heilongjiang Province. Scientia Agricultura Sinica, 2022, 55(4): 625-640. (in Chinese) | |
| [2] |
ZHANG Y L, ZHU Q L, YAO Y X, ZHAO Z H, CORRELL J C, WANG L, PAN Q H. The race structure of the rice blast pathogen across southern and northeastern China. Rice, 2017, 10(1): 46.
doi: 10.1186/s12284-017-0185-y pmid: 28983868 |
| [3] | 孟峰, 张亚玲, 靳学慧, 张晓玉, 姜军. 黑龙江省稻瘟病菌无毒基因AVR-Pib、AVR-Pik和AvrPiz-t的检测与分析. 中国农业科学, 2019, 52(23): 4262-4273. |
| MENG F, ZHANG Y L, JIN X H, ZHANG X Y, JIANG J. Detection and analysis of Magnaporthe oryzae avirulence genes AVR-Pib, AVR-Pik and AvrPiz-t in Heilongjiang Province. Scientia Agricultura Sinica, 2019, 52(23): 4262-4273. (in Chinese) | |
| [4] |
FLOR H H. Current status of the gene-for-gene concept. Annual Review of Phytopathology, 1971, 9: 275-296.
doi: 10.1146/annurev.py.09.090171.001423 |
| [5] | 吴伟怀, 王玲, 程贯忠, 朱有勇, 潘庆华. 稻瘟病菌群体的分子遗传学研究——广东省与云南省稻瘟病菌群体遗传及致病型结构的比较分析. 中国农业科学, 2004, 37(5): 675-680. |
| WU W H, WANG L, CHENG G Z, ZHU Y Y, PAN Q H. Studies on molecular genetics of rice blast fungus population——Comparison of genetic and pathotypic structures of two rice blast fungus populations derived from Guangdong and Yunnan provinces of China. Scientia Agricultura Sinica, 2004, 37(5): 675-680. (in Chinese) | |
| [6] |
ORBACH M J, FARRALL L, SWEIGARD J A, CHUMLEY F G, VALENT B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. The Plant Cell, 2000, 12(11): 2019-2032.
doi: 10.1105/tpc.12.11.2019 |
| [7] |
COLLEMARE J, PIANFETTI M, HOULLE A E, MORIN D, CAMBORDE L, GAGEY M J, BARBISAN C, FUDAL I, LEBRUN M H, BOHNERT H U. Magnaporthe grisea avirulence gene ACE1 belongs to an infection-specific gene cluster involved in secondary metabolism. New Phytologist, 2008, 179(1): 196-208.
doi: 10.1111/j.1469-8137.2008.02459.x |
| [8] |
FARMAN M L, LEONG S A. Chromosome walking to the AVR1-CO39 avirulence gene of Magnaporthe grisea: Discrepancy between the physical and genetic maps. Genetics, 1998, 150(3): 1049-1058.
doi: 10.1093/genetics/150.3.1049 |
| [9] |
LI W, WANG B H, WU J, LU G D, HU Y J, ZHANG X, ZHANG Z G, ZHAO Q, FENG Q, ZHANG H Y, WANG Z Y, WANG G L, HAN B, WANG Z H, ZHOU B. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Molecular Plant-Microbe Interactions, 2009, 22(4): 411-420.
doi: 10.1094/MPMI-22-4-0411 |
| [10] |
YOSHIDA K, SAITOH H, FUJISAWA S, KANZAKI H, MATSUMURA H, YOSHIDA K, TOSA Y, CHUMA I, TAKANO Y, WIN J, KAMOUN S, TERAUCHI R. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. The Plant Cell, 2009, 21(5): 1573-1591.
doi: 10.1105/tpc.109.066324 |
| [11] |
WU J, KOU Y J, BAO J D, LI Y, TANG M Z, ZHU X L, PONAYA A, XIAO G, LI J B, LI C Y, et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9that triggers Pi9- mediated blast resistance in rice. New Phytologist, 2015, 206(4): 1463-1475.
doi: 10.1111/nph.13310 |
| [12] |
ZHANG S L, WANG L, WU W H, HE L Y, YANG X F, PAN Q H. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Scientific Reports, 2015, 5: 11642.
doi: 10.1038/srep11642 |
| [13] |
KANG S, SWEIGARD J A, VALENT B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Molecular Plant- Microbe Interactions, 1995, 8(6): 939-948.
doi: 10.1094/MPMI-8-0939 |
| [14] | SWEIGARD J A, CARROLL A M, KANG S, FARRALL L, CHUNLEY F G, VALENT B. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. The Plant Cell, 1995, 7(8): 1221-1233. |
| [15] | MASAKI H I. Cloning and characterization of PWL1 and PWL2 host-species specificity genes of the finger millet blast pathogen, Magnaporthe oryzae[D]. Kenya: Pwani University, 2020. |
| [16] | 余欢, 姜华, 王艳丽, 孙国昌. 无毒基因在不同寄主梨孢菌中的变异研究. 浙江农业学报, 2015, 27(8): 1414-1421. |
| YU H, JIANG H, WANG Y L, SUN G C. Variability of avirulence genes in Pyricularia isolates from different hosts. Acta Agriculturae Zhejiangensis, 2015, 27(8): 1414-1421. (in Chinese) | |
| [17] | 穆慧敏. 利用无毒基因研究浙江省分离于不同年份的稻瘟病菌群体的遗传结构多样性[D]. 南京: 南京农业大学, 2013. |
| MU H M. Analysis of genetic diversity of Magnaporthe oryzae isolates from Zhejiang Province, over the past thirty years based on avirulence gene[D]. Nanjing: Nanjing Agricultural University, 2013. (in Chinese) | |
| [18] | 孟峰, 靳学慧, 张亚玲. PWL基因家族在黑龙江省水稻稻瘟病菌中的分布与变异. 植物保护, 2020, 46(6): 71-76. |
| MENG F, JIN X H, ZHANG Y L. Distribution and variability of Magnaporthe oryzae PWL gene family in Heilongjiang Province. Plant Protection, 2020, 46(6): 71-76. (in Chinese) | |
| [19] | 李焕宇, 付婷婷, 张云, 吕天佑, 李远, 徐秉良. 5种方法提取真菌基因组DNA作为PCR模板效果的比较. 中国农学通报, 2017, 33(16): 28-35. |
| LI H Y, FU T T, ZHANG Y, LÜ T Y, LI Y, XU B L. Effect comparison of five methods to extract fungal genomic DNA as PCR templates. Chinese Agricultural Science Bulletin, 2017, 33(16): 28-35. (in Chinese) | |
| [20] | 聂江山. 稻瘟病菌群体致病型结构的动态变化及其区域特征[D]. 广州: 华南农业大学, 2017. |
| NIE J S. Temporal dynamics and spatial variation of pathotype structure of Magnaporthe oryzae populations in China[D]. Guangzhou: South China Agricultural University, 2017. (in Chinese) | |
| [21] | 穆慧敏, 姜华, 毛雪琴, 柴荣耀, 王艳丽, 张震, 王教瑜, 邱海萍, 杜新法, 孙国昌. PWL基因家族在稻瘟病菌中的分布及变异初探. 浙江农业学报, 2013, 25(3): 526-532. |
| MU H M, JIANG H, MAO X Q, CHAI R Y, WANG Y L, ZHANG Z, WANG J Y, QIU H P, DU X F, SUN G C. A primary study on distribution and variability of PWL gene family in Magnaporthe grisea. Acta Agriculturae Zhejiangensis, 2013, 25(3): 526-532. (in Chinese) | |
| [22] |
SHI N N, RUAN H C, LIU X Z, YANG X J, DAI Y L, GAN L, CHEN F R, DU Y X. Virulence structure of Magnaporthe oryzae populations from Fujian Province, China. Canadian Journal of Plant Pathology, 2018, 40(4): 542-550.
doi: 10.1080/07060661.2018.1504821 |
| [23] | 张崎峰, 靳学慧, 蔡鑫鑫, 李金良, 陈海军. 黑龙江省稻瘟病菌无毒基因的检测. 黑龙江农业科学, 2014(12): 70-73. |
| ZHANG Q F, JIN X H, CAI X X, LI J L, CHEN H J. Detection on avirulence gene of rice blast in Heilongjiang Province. Heilongjiang Agricultural Sciences, 2014(12): 70-73. (in Chinese) | |
| [24] | 曹雪琦. 福建省稻瘟病菌田间种群无毒基因的变异检测[D]. 福州: 福建农林大学, 2020. |
| CAO X Q. Detection of avirulence gene mutations in the field population of rice blast flungus in Fujian Province[D]. Fuzhou: Fujian Agriculture and Forestry University, 2020. (in Chinese) | |
| [25] |
ZEIGLER R S, CUOC L X, SCOTT R P, BERNARDO M A, CHEN D H, VALENT B, NELSON R J. The relationship between lineage and virulence in Pyricularia grisea in the Philippines. Phytopathology, 1995, 85(4): 443-451.
doi: 10.1094/Phyto-85-443 |
| [26] |
GOMEZ LUCIANO L B, TSAI I J, CHUMA I, TOSA Y, CHEN Y H, LI J Y, LI M Y, LU M Y J, NAKAYASHIKI H, LI W H. Blast fungal genomes show frequent chromosomal changes, gene gains and losses, and effector gene turnover. Molecular Biology and Evolution, 2019, 36(6): 1148-1161.
doi: 10.1093/molbev/msz045 pmid: 30835262 |
| [27] |
YOSHIDA K, SAUNDERS D G, MITSUOKA C, NATSUME S, KOSUGI S, SAITOH H, INOUE Y, CHUMA I, TOSA Y, CANO L M, KAMOUN S, TERAUCHI R. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics, 2016, 17: 370.
doi: 10.1186/s12864-016-2690-6 |
| [28] |
BAO J, CHEN M, ZHONG Z, TANG W, LIN L, ZHANG X, JIANG H, ZHANG D, MIAO C, TANG H, et al. PacBio sequencing reveals transposable elements as a key contributor to genomic plasticity and virulence variation in Magnaporthe oryzae. Molecular Plant, 2017, 10(11): 1465-1468.
doi: 10.1016/j.molp.2017.08.008 |
| [29] |
THON M R, PAN H, DIENER S, PAPALAS J, TARO A, MITCHELL T K, DEAN R A. The role of transposable element clusters in genome evolution and loss of synteny in the rice blast fungus Magnaporthe oryzae. Genome Biology, 2006, 7(2): R16.
doi: 10.1186/gb-2006-7-2-r16 |
| [30] |
CHUMA I, ISOBE C, HOTTA Y, IBARAGI K, FUTAMATA N, KUSABA M, YOSHIDA K, TERAUCHI R, FUJITA Y, NAKAYASHIKI H, VALENT B, TOSA Y. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathogens, 2011, 7(7): e1002147.
doi: 10.1371/journal.ppat.1002147 |
| [31] |
SCHNEIDER D R S, SARAIVA A M, AZZONI A R, MIRANDA H R C, DE TOLEDO M A S, PELLOSO A C, SOUZA A P. Overexpression and purification of PWL2D, a mutant of the effector protein PWL2 from Magnaporthe grisea. Protein Expression and Purification, 2010, 74(1): 24-31.
doi: 10.1016/j.pep.2010.04.020 |
| [32] |
申浩冉, 白辉, 任世龙, 王璐, 王永芳, 全建章, 董志平, 李志勇. 谷瘟病菌无毒基因PWL家族检测及变异分析. 华北农学报, 2020, 35(3): 178-183.
doi: 10.7668/hbnxb.20190568 |
|
SHEN H R, BAI H, REN S L, WANG L, WANG Y F, QUAN J Z, DONG Z P, LI Z Y. Detection and variation analysis of PWL gene family of Magnaporthe oryzae in foxtail millet. Acta Agriculturae Boreali-Sinica, 2020, 35(3): 178-183. (in Chinese)
doi: 10.7668/hbnxb.20190568 |
| [1] | 汪文娟,苏菁,陈深,杨健源,陈凯玲,冯爱卿,汪聪颖,封金奇,陈炳,朱小源. 广东省侵染美香占2号的稻瘟病菌致病性及无毒基因变异分析[J]. 中国农业科学, 2022, 55(7): 1346-1358. |
| [2] | 郭燕, 张树航, 李颖, 张馨方, 王广鹏. 中国板栗36个叶片表型性状的多样性[J]. 中国农业科学, 2022, 55(5): 991-1009. |
| [3] | 黄勋和,翁茁先,李威娜,王庆,何丹林,罗威,张细权,杜炳旺. 中国地方品种黄鸡线粒体DNA D-loop遗传多样性研究[J]. 中国农业科学, 2022, 55(22): 4526-4538. |
| [4] | 万映伶,朱梦婷,刘爱青,金亦佳,刘燕. 中国观赏芍药表型多样性解析与资源评价[J]. 中国农业科学, 2022, 55(18): 3629-3639. |
| [5] | 郭迎新,陈永亮,苗琪,范志勇,孙军伟,崔振岭,李军营. 洱海流域植烟土壤养分时空变异特征及肥力评价[J]. 中国农业科学, 2022, 55(10): 1987-1999. |
| [6] | 吴云雨,肖宁,余玲,蔡跃,潘存红,李育红,张小祥,黄年生,季红娟,戴正元,李爱宏. 长江下游粳稻稻瘟病广谱抗性基因组合模式分析[J]. 中国农业科学, 2021, 54(9): 1881-1893. |
| [7] | 陈璨,韩南南,刘洋,史晓维,司红起,马传喜. 小麦Glu-3位点基因拷贝数的变异分析[J]. 中国农业科学, 2021, 54(6): 1092-1103. |
| [8] | 李紫腾,曹钰晗,李楠,孟祥龙,胡同乐,王树桐,王亚南,曹克强. 苹果锈果类病毒在7个品种苹果上的分子变异及系统发育关系[J]. 中国农业科学, 2021, 54(20): 4326-4336. |
| [9] | 刘有春,刘威生,王兴东,孙斌,刘修丽,杨艳敏,魏鑫,杨玉春,张舵,刘成,李天忠. 基于简化基因组测序的越橘杂交后代鉴定[J]. 中国农业科学, 2021, 54(2): 370-378. |
| [10] | 张林林,智慧,汤沙,张仁梁,张伟,贾冠清,刁现民. 谷子抽穗时间基因SiTOC1的表达与单倍型变异分析[J]. 中国农业科学, 2021, 54(11): 2273-2286. |
| [11] | 盖钧镒,贺建波. 限制性两阶段多位点全基因组关联分析法(RTM-GWAS)的特点、常见提问与应用前景[J]. 中国农业科学, 2020, 53(9): 1699-1703. |
| [12] | 贺建波,刘方东,王吴彬,邢光南,管荣展,盖钧镒. 限制性两阶段多位点全基因组关联分析法在遗传育种中的应用[J]. 中国农业科学, 2020, 53(9): 1704-1716. |
| [13] | 普雪可,吴春花,勉有明,苗芳芳,侯贤清,李荣. 不同覆盖方式对旱作马铃薯生长及土壤水热特征的影响[J]. 中国农业科学, 2020, 53(4): 734-747. |
| [14] | 赵晴月,许世杰,张务帅,张哲,姚智,陈新平,邹春琴. 中国玉米主产区土壤养分的空间变异及影响因素分析[J]. 中国农业科学, 2020, 53(15): 3120-3133. |
| [15] | 秦艳红,王永江,王爽,乔奇,田雨婷,张德胜,张振臣. 甘薯羽状斑驳病毒O株系和RC株系中国分离物全基因组 序列分析及其遗传特征[J]. 中国农业科学, 2020, 53(11): 2207-2218. |
|
||