













中国农业科学 ›› 2026, Vol. 59 ›› Issue (4): 793-806.doi: 10.3864/j.issn.0578-1752.2026.04.007
江峰1( ), 吴春燕1, 王亦好1, 杨泽众2, 龚成1(
), 吴春燕1, 王亦好1, 杨泽众2, 龚成1( ), 罗晨1
), 罗晨1
收稿日期:2025-11-15
出版日期:2026-02-10
发布日期:2026-02-10
通信作者:
联系方式:
江峰,E-mail:jiangfeng23333@163.com。
基金资助:
JIANG Feng1( ), WU ChunYan1, WANG YiHao1, YANG ZeZhong2, GONG Cheng1(
), WU ChunYan1, WANG YiHao1, YANG ZeZhong2, GONG Cheng1( ), LUO Chen1
), LUO Chen1
Received:2025-11-15
Published:2026-02-10
Online:2026-02-10
摘要:
【目的】烟粉虱(Bemisia tabaci)的宿主适应性与其脂类物质代谢(特别是脂肪酸代谢)的可塑性密切相关。脂肪酸延长酶(fatty acid elongase,ELO)为脂肪酸代谢网络的核心调控因子,系统解析其基因家族,揭示烟粉虱营养适应与繁殖发育的代谢调控机制,为害虫绿色防控机制提供新型靶标基因资源。【方法】基于烟粉虱MED基因组和其他昆虫的ELO基因序列,通过多重序列比对,鉴定烟粉虱ELO,采用RT-PCR扩增并克隆获得22条BtELO基因序列。利用生物信息学工具系统分析其基因结构、蛋白理化性质,利用最大似然法构建BtELO与其他半翅目昆虫ELO的系统进化树。通过实时荧光定量PCR(qRT-PCR)方法检测BtELO在烟粉虱不同发育阶段(卵、1—4龄若虫、成虫)及成虫不同组织(头、胸、腹)中的时空表达谱。【结果】成功鉴定并克隆得到22条BtELO基因CDS序列,长度为714—1 296 bp,分布在烟粉虱MED基因组10条SCAFFOLD上,按照线性排列顺序依次命名为BtELO1—22,编码237—431个氨基酸,蛋白整体呈现出疏水性,含有6—7个跨膜结构域,亚细胞定位分析预测均在内质网中;氨基酸序列比对和系统进化分析显示,BtELO蛋白具有典型的ELO保守结构域和组氨酸簇(HXXHH),BtELO基因家族内部成员相似度最高达到76%,BtELO可分为7大进化分支,并呈现出较好的系统发育聚类特征,进化关系保守,部分BtELO形成相对独立分支,存在基因复制事件;时空表达谱分析显示,BtELO在烟粉虱的不同发育阶段和组织中的表达具有明显时空特异性,表明BtELO在烟粉虱中可能发挥着多种功能。【结论】烟粉虱22条BtELO含有典型的延长酶结构域及HXXHH保守基序,其在烟粉虱不同发育阶段和组织中均有表达,部分基因在卵期和成虫腹部高表达,推测其在生殖过程中发挥作用,而在若虫期高表达的基因可能与生长发育及蜕皮相关。
江峰, 吴春燕, 王亦好, 杨泽众, 龚成, 罗晨. 烟粉虱MED脂肪酸延长酶基因家族鉴定和表达分析[J]. 中国农业科学, 2026, 59(4): 793-806.
JIANG Feng, WU ChunYan, WANG YiHao, YANG ZeZhong, GONG Cheng, LUO Chen. Identification and Expression Analysis of the Fatty Acid Elongase Gene Family in Bemisia tabaci MED[J]. Scientia Agricultura Sinica, 2026, 59(4): 793-806.
表1
引物序列"
| 基因 Gene | 登录号 Accession number | 引物序列Primer sequence (5′-3′) | |
|---|---|---|---|
| 开放阅读框扩增Amplification of ORF | 实时荧光定量PCR qRT-PCR | ||
| BtELO1 | PX527230 | ATGGCAACCGAAGTAGTTTTAG | CTGGGACTGCTTATCTTCAACTGT |
| GGATCATTTCATTTTATTTTCGTCA | CAGGAAAAAGAATGACGAACAATC | ||
| BtELO2 | PX527231 | AATGCTGCTGGTGGTAGTGAA | GTGTGGTGATATACATGGAGAA |
| TCGAGTAGGCTTCAAGGTAACAT | AACGCCGGAGAACATTAC | ||
| BtELO3 | PX527232 | CACCTTGGATCTTGGATAGTTT | GGGCCATGGAATGAGTTTAG |
| GTACGAGAAGGTAACCTGTGCT | TCCAATTCGACCTCTTGTATTC | ||
| BtELO4 | PX527233 | TATTGATTTTGGATCGCCGA | GTAGGCCCACGTTGATAAG |
| GAGCGTTGCTTGTCCGTAAA | GTGTTCTTCGTACTCCGTAAA | ||
| BtELO5 | PX527234 | CGAATCGTTGGTGAACTGAC | ATACTCTTCGCCGACTTCT |
| AGAGCGGAGGTCAACGACTA | CGAACAGTCTAGGATAGCTTTG | ||
| BtELO6 | PX527235 | TCCGTATAACCACATCTCAGGAC | CATGAATCAAGGCGTTCAATAC |
| AAACAAGATTACAACTGGAATTCA | CCTCCACGTTTATCACCATT | ||
| BtELO7 | PX527236 | TCCCCGGATTTTTATTAATTTGT | TGGAACTACCTCATGTTGAAAG |
| CAGTTGAATGAATTGACAGCGAT | ACCATCTGAGTGTGATGATAGA | ||
| BtELO8 | PX527237 | TCCAGCTCCGCATGATTG | GAGCCGATGTGAAATGCTGATC |
| ATGTGTGGATCGCCAATAGATAG | GCTCCCACCTTTTTACTTTTGCTA | ||
| BtELO9 | PX527238 | ATGACGCAGATTCAAAGGGG | ACATGACTCAACTCCAAATAGGTCA |
| TTTCAACGGAGATCATGTGGTT | AAAACGAAGATGTTCACTGCGAT | ||
| BtELO10 | PX527239 | ATGGCGCAGATTCAAAGACAA | TTGTTCATATCTGGATGTACGGGTA |
| CGAAATTGCATTGCGATCC | CTGATTTGGAGTTGAGTGATGTGTT | ||
| BtELO11 | PX527240 | CCTTCCGACTCCAATATACCTT | GAGTTCTTCGACACCTTCTTC |
| GCAAGTTCCTCGCAATCAT | CATGATACCGTGGTGGATAAC | ||
| BtELO12 | PX527241 | AGGGAAGCCTATTTGACGTTAA | TCTTTCAGAATGGTTGGCTCG |
| GCTCTTCTCCCGTCCAGATT | TGTTTCTCCCAAGAGGCGTG | ||
| BtELO13 | PX527242 | TAAAATAGTGTCTTGCTTATCGTCT | AGCGACCATGTAGTAGAAGTA |
| TTCACCCAGCTCAGATAACTATG | GTGTCTGGATGGGAATGAAA | ||
| BtELO14 | PX527243 | TTTATTTGTCGGTCCTACTCATT | CTGAAGAGCCAGAAGAAGAAG |
| CTGTACAGAGGAATCACGACTT | CTCCTGTCAACTGCTTATCTAC | ||
| BtELO15 | PX527244 | GTTTACGCTGGTGAAGTGGACT | GCTGTGGATAGGTACCATTATT |
| ACTAAGAAAGGGCTCTGTGACG | TTGCCAAAGTGGGAATAGG | ||
| BtELO16 | PX527245 | CTGGAATGATTCGCTGGC | GGACGACCAAGTTCCATTTA |
| TTTTCAACGGGCATCTTCA | TGCGAGCTCTGCTATCT | ||
| BtELO17 | PX527246 | CCTCTCGTACCCACACTTACC | CTCTCACCAACTACTCCTACA |
| AAGAGAGTTCAACGCAGTCAAT | CATGTAGAGTCCGCAATAGTAAA | ||
| BtELO18 | PX527247 | ATGTTCAAACGGGTTCTCTGC | ATCATAGTCTGCGTGCTGTTCTTC |
| CTTTTACAAACAATAGCCAGTCCA | CTTTGTCTTCTTATCGCTGGTGAG | ||
| BtELO19 | PX527248 | ATTCGGGCTCACTTGTGTG | AAGAACGTGTCGCAGAAC |
| AAGTATCATCACGAGGAAAAATC | GCCGGTGACTTTCGTAAATA | ||
| BtELO20 | PX527249 | CCAAGTGATTCTCTAACCTATTGATT | GTCCGAGAAGAGGAAGTAGA |
| GTCATACGCCCTTTTTGCC | ATCCACGCTTTCCAACTAC | ||
| BtELO21 | PX527250 | ATCCGACTTCGTGCAGGGAT | GGGAGAGACCATTTGCATAG |
| TATTCCTTTCTTTTCAGGGCTC | GCTCATGCAACCACAAATG | ||
| BtELO22 | PX527251 | GGCTCGTCATACAGACCCAA | CTTCACGTCTATCACCACAC |
| TTGGAATTTGCGGAGACACT | CTGAATTGATCACGCCGATA | ||
| EF1α | EE600682 | — | TAGCCTTGTGCCAATTTCCG |
| — | CCTTCAGCATTACCGTCC | ||
表2
烟粉虱ELO蛋白理化性质分析"
| 蛋白 Protein | 氨基酸数量 Amino acids size (aa) | 分子量 Mw (kDa) | 等电点 pI | 不稳定系数 Instability index (II) | 脂肪系数 Aliphatic index | 亲水性平均系数 Grand of hydropathicity |
|---|---|---|---|---|---|---|
| BtELO1 | 272 | 31.93 | 9.16 | 43.10 | 100.33 | 0.424 |
| BtELO2 | 338 | 39.34 | 9.40 | 39.16 | 92.19 | -0.086 |
| BtELO3 | 270 | 31.90 | 9.37 | 30.44 | 100.33 | 0.239 |
| BtELO4 | 289 | 33.66 | 9.44 | 31.90 | 108.24 | 0.348 |
| BtELO5 | 268 | 31.59 | 9.57 | 35.59 | 104.70 | 0.390 |
| BtELO6 | 290 | 33.74 | 9.66 | 35.21 | 104.79 | 0.379 |
| BtELO7 | 276 | 32.54 | 9.58 | 25.79 | 105.18 | 0.362 |
| BtELO8 | 277 | 32.62 | 9.59 | 32.23 | 106.53 | 0.348 |
| BtELO9 | 280 | 33.26 | 9.70 | 31.28 | 111.00 | 0.394 |
| BtELO10 | 277 | 33.09 | 9.56 | 38.26 | 106.97 | 0.306 |
| BtELO11 | 313 | 36.29 | 9.53 | 33.65 | 84.76 | 0.157 |
| BtELO12 | 277 | 32.62 | 9.66 | 32.16 | 104.08 | 0.353 |
| BtELO13 | 343 | 39.98 | 9.48 | 44.96 | 69.62 | -0.049 |
| BtELO14 | 333 | 38.48 | 9.55 | 34.40 | 79.64 | -0.075 |
| BtELO15 | 431 | 50.02 | 9.02 | 24.04 | 78.47 | -0.309 |
| BtELO16 | 269 | 31.04 | 9.31 | 41.58 | 112.71 | 0.380 |
| BtELO17 | 292 | 34.44 | 9.41 | 48.91 | 82.81 | 0.121 |
| BtELO18 | 289 | 33.63 | 9.37 | 24.19 | 91.45 | 0.242 |
| BtELO19 | 301 | 35.38 | 8.92 | 42.34 | 95.55 | 0.123 |
| BtELO20 | 322 | 37.72 | 9.44 | 29.24 | 78.11 | -0.065 |
| BtELO21 | 319 | 37.35 | 9.57 | 35.48 | 83.98 | 0.096 |
| BtELO22 | 237 | 27.90 | 9.52 | 26.00 | 106.88 | 0.387 |

图4
烟粉虱、褐飞虱与黑腹果蝇ELO蛋白的多序列比对 NlELO10:褐飞虱N. lugens (AWJ25044.1);bond:黑腹果蝇D. melanogaster (NP_651062.3)。黑色区域Black area:相似度100% 100% similarity;粉红色区域Pink area:相似度大于75% Similarity greater than 75%;浅蓝色区域Light blue area:相似度大于50% Similarity greater than 50%;红色虚线Red dashed line:ELO结构域ELO domain;蓝色横线Blue horizontal line:组氨酸簇(HXXHH)Histidine cluster (HXXHH)"

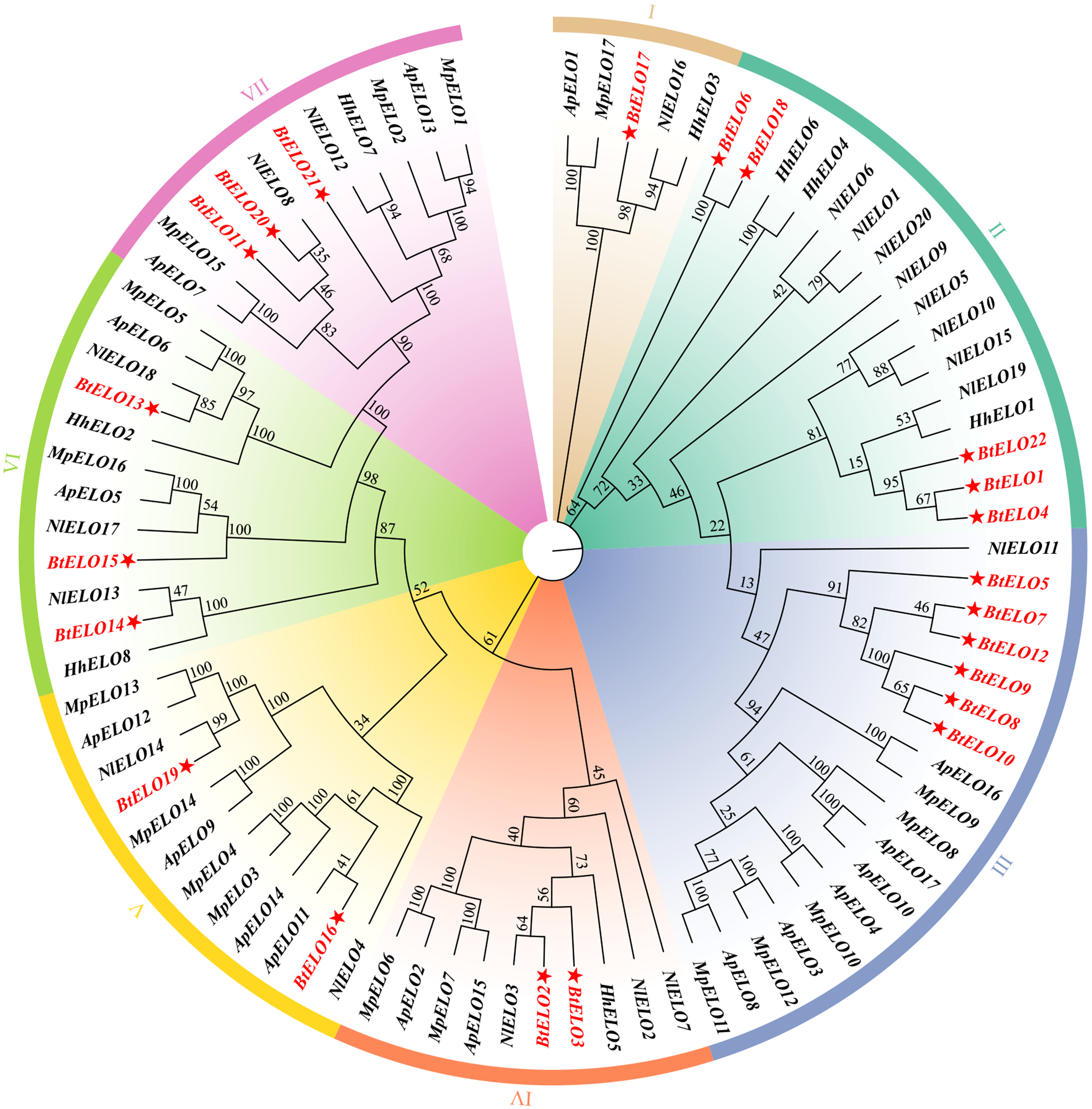
图5
BtELO与其他半翅目昆虫ELO的系统进化树 ELO蛋白来源物种及GenBank登录号Origin species and their GenBank accession numbers褐飞虱N. lugens:NlELO1-NlELO20 (AWJ25035.1, AWJ25036.1, WJ25037.1, AWJ25038.1, AWJ25039.1, AWJ25040.1, AWJ25041.1, AWJ25042.1, AWJ25043.1, AWJ25044.1, AWJ25045.1, AWJ25046.1, AWJ25047.1, AWJ25048.1, AWJ25049.1, AWJ25050.1, AWJ25051.1, AWJ25052.1, AWJ25053.1, AWJ25054.1);豌豆蚜A. pisum:ApELO1-ApELO17 (NP_001156725.1, NP_001280394.1, NP_001280432.1, XP_001942907.1, XP_001950768.5, XP_001952817.1, XP_001952818.2, XP_003240836.1, XP_003245187.1, XP_003247564.1, XP_008182500.2, XP_008184040.1, XP_008185301.1, XP_016659383.1, XP_029341901.1, XP_029342443.1, XP_029342445.1);桃蚜M. persicae:MpELO1-MpELO17 (XP_022169899.1, XP_022169905.1, XP_022172237.1, XP_022172238.1, XP_022173786.1, XP_022174580.1, XP_022174581.1, XP_022177768.1, XP_022177770.1, XP_022177781.1, XP_022177785.1, XP_022177786.1, XP_022178544.1, XP_022178610.1, XP_022178694.1, XP_022181274.1, XP_022183411.1);茶翅蝽H. halys:HhELO1-HhELO8 (XP_014274939.1, XP_014276092.1, XP_014276467.1, XP_024215601.2, XP_024217007.2, XP_066901701.1, XP_066902872.1, XP_066905193.1)"

| [1] |
doi: 10.1016/j.cell.2021.02.014 pmid: 33770502 |
| [2] |
doi: 10.1016/j.cropro.2016.11.035 |
| [3] |
doi: 10.1016/S0261-2194(01)00108-9 |
| [4] |
|
| [5] |
郑天祥, 钱雨农, 张大羽. 昆虫脂肪酸合成通路关键基因的研究进展. 中国生物工程杂志, 2017, 37(11): 19-27.
|
|
|
|
| [6] |
pmid: 14636670 |
| [7] |
doi: 10.1038/s41594-021-00609-2 |
| [8] |
doi: 10.1016/j.plipres.2006.01.004 pmid: 16564093 |
| [9] |
doi: 10.1016/j.bbrc.2005.06.015 |
| [10] |
doi: 10.1038/ncomms9263 |
| [11] |
doi: 10.1016/j.ibmb.2019.03.005 |
| [12] |
doi: 10.1002/arch.v119.1 |
| [13] |
doi: 10.1016/j.jinsphys.2020.104052 |
| [14] |
doi: 10.1021/acs.jafc.3c07412 |
| [15] |
姚晶, 郭晓军, 王甦, 王昱超, 罗晨, 张帆, 李绍勤. 利用TaqMan等位基因技术鉴定烟粉虱MEAM1和MED隐种. 昆虫学报, 2013, 56(1): 98-103.
|
|
|
|
| [16] |
doi: 10.1371/journal.pone.0053006 |
| [17] |
|
| [18] |
doi: 10.1186/s40001-023-01523-7 pmid: 37981715 |
| [19] |
doi: 10.1139/gen-2017-0224 |
| [20] |
doi: 10.1242/jcs.006551 |
| [21] |
|
| [22] |
doi: 10.1111/ins.v30.1 |
| [23] |
doi: 10.1016/j.ibmb.2015.03.003 |
| [24] |
doi: 10.3390/ijms25116080 |
| [25] |
doi: 10.1093/plcell/koac076 pmid: 35253876 |
| [26] |
王宏燕. 柑橘全爪螨对螺螨双酯胁迫的响应及其分子机制[D]. 南昌: 南昌大学, 2024.
|
|
|
|
| [27] |
|
| [28] |
|
| [29] |
doi: 10.1371/journal.pgen.1011186 |
| [30] |
|
| [31] |
doi: 10.1038/s41437-020-00380-y pmid: 33139902 |
| [32] |
doi: 10.1002/bies.201500014 pmid: 25988392 |
| [33] |
doi: S2214-5745(17)30119-0 pmid: 30025637 |
| [34] |
doi: 10.3389/fphys.2019.00913 |
| [35] |
doi: 10.1016/j.ibmb.2016.10.011 |
| [1] | 罗正英, 胡嗣桢, 林秀琴, 胡鑫, 张敏, 徐超华, 刘新龙, 曾千春. 甘蔗属割手密与热带种PEBP基因家族的鉴定及其开花调控功能分析[J]. 中国农业科学, 2026, 59(4): 734-749. |
| [2] | 吴琼, 谢香庭, 王磊, 牟勇, 李进伟. 转基因大豆DBN8205转化体特异性定量PCR方法的研发和验证[J]. 中国农业科学, 2026, 59(1): 29-40. |
| [3] | 杨彩丽, 李永洲, 贺亮亮, 宋银花, 章鹏, 刘肇先, 李鹏慧, 刘三军. 葡萄TPS基因家族全基因组鉴定及VvTPS4在单萜形成中的功能验证[J]. 中国农业科学, 2025, 58(7): 1397-1417. |
| [4] | 张天雨, 李白, 藏金萍, 曹宏哲, 董金皋, 邢继红, 张康. 灰葡萄孢HMG家族基因的全基因组鉴定与表达规律分析[J]. 中国农业科学, 2025, 58(4): 704-718. |
| [5] | 李建昆, 彭超, 张子扬, 梁茜, 魏鸣康, 杨强, 李斌奇, Muhammad Moaaz Ali, Viola Kayima, 陈发兴, 邓红红. ‘皇冠’李果实空腔褐变与木质素积累的关系及其4CL基因家族鉴定[J]. 中国农业科学, 2025, 58(4): 759-778. |
| [6] | 仪泽会, 王颖, 宋慧霞, 赵婧, 毛丽萍. 芦笋过氧化物还原酶(Peroxiredoxins)基因家族全基因组鉴定及表达分析[J]. 中国农业科学, 2025, 58(18): 3728-3743. |
| [7] | 齐香玉, 李新茹, 陈双双, 冯景, 陈慧杰, 刘欣童, 金玉妍, 邓衍明. 茉莉花FLA基因家族鉴定及JsFLA2的功能分析[J]. 中国农业科学, 2025, 58(17): 3516-3530. |
| [8] | 王伟, 吴传磊, 胡晓渝, 李佳佳, 白鹏宇, 王郭伋, 苗龙, 王晓波. 大豆LOX基因家族的全基因组鉴定及GmLOX15A1基因等位变异对百粒重的影响[J]. 中国农业科学, 2025, 58(1): 10-29. |
| [9] | 王程泽, 张燕, 付伟, 贾京哲, 董金皋, 申珅, 郝志敏. 玉米ACO基因家族生物信息学及表达模式分析[J]. 中国农业科学, 2024, 57(7): 1308-1318. |
| [10] | 曾祥翠, 杨永念, 李如月, 蒋学乾, 蒋旭, 徐嫣然, 刘忠宽, 龙瑞才, 康俊梅, 杨青川, 李明娜. 紫花苜蓿MsCEP基因家族的鉴定及其调控根系生长发育功能的分析[J]. 中国农业科学, 2024, 57(24): 4839-4853. |
| [11] | 赵婕, 赵龙缘, 潘凝辉, 管丽蓉, 杜云龙, 李成云, 王云月, 谢勇. 水解酶基因BGIOSGA023826在稻瘟菌侵染过程中的抗病表型效应[J]. 中国农业科学, 2024, 57(23): 4607-4618. |
| [12] | 吴传磊, 胡晓渝, 王伟, 苗龙, 白鹏宇, 王郭伋, 李娜, 舒阔, 邱丽娟, 王晓波. 大豆油分相关功能基因分子标记开发与鉴定及优异等位变异聚合分析[J]. 中国农业科学, 2024, 57(22): 4402-4415. |
| [13] | 钟梓春, 吴洪鑫, 张杰, 郭玉静, 何柳燕, 许小霞, 金丰良, 庞锐. 三种稻飞虱Toll受体基因家族比较分析[J]. 中国农业科学, 2024, 57(20): 4007-4021. |
| [14] | 孙思思, 马伍, 司惠茹, 王贤忠, 柳强, 罗炎琳, 陈孝玉龙, 唐斌. 豌豆修尾蚜水通道蛋白特征及响应高湿度胁迫表达变化[J]. 中国农业科学, 2024, 57(20): 4057-4070. |
| [15] | 尹军良, 李婧怡, 韩硕, 杨培华, 马佳伟, 刘奕清, 胡海骏, 朱永兴. 生姜NHX基因家族成员鉴定及其在硅缓解盐胁迫中的表达特征[J]. 中国农业科学, 2024, 57(19): 3848-3869. |
|
||