













中国农业科学 ›› 2022, Vol. 55 ›› Issue (7): 1346-1358.doi: 10.3864/j.issn.0578-1752.2022.07.007
汪文娟( ),苏菁,陈深,杨健源,陈凯玲,冯爱卿,汪聪颖,封金奇,陈炳,朱小源(
),苏菁,陈深,杨健源,陈凯玲,冯爱卿,汪聪颖,封金奇,陈炳,朱小源( )
)
收稿日期:2021-11-01
接受日期:2021-12-20
出版日期:2022-04-01
发布日期:2022-04-18
通讯作者:
朱小源
作者简介:汪文娟,E-mail: 基金资助:
WANG WenJuan( ),SU Jing,CHEN Shen,YANG JianYuan,CHEN KaiLing,FENG AiQing,WANG CongYing,FENG JinQi,CHEN Bing,ZHU XiaoYuan(
),SU Jing,CHEN Shen,YANG JianYuan,CHEN KaiLing,FENG AiQing,WANG CongYing,FENG JinQi,CHEN Bing,ZHU XiaoYuan( )
)
Received:2021-11-01
Accepted:2021-12-20
Online:2022-04-01
Published:2022-04-18
Contact:
XiaoYuan ZHU
摘要:
【目的】分析侵染广东稻区优质品种美香占2号稻瘟病菌(Magnaporthe oryzae)的致病性及无毒基因变异特征,为美香占2号在不同稻区的合理布局提供参考依据。【方法】利用9个抗稻瘟病单基因系,对稻瘟病菌单孢分离菌株进行致病性测定;并利用8个稻瘟病菌已克隆无毒基因的功能性分子标记对2013—2018年不同年份、不同稻区采集的稻瘟病菌单孢分离菌株的DNA进行PCR扩增,通过琼脂糖凝胶电泳检测分析,选取不同带型和不同年份代表性菌株的相应无毒基因全长或CDS区域的PCR片段进行测序。测序结果与相应无毒基因的参考序列进行碱基序列的比较分析;并利用相应的水稻抗稻瘟病单基因系,对无毒基因不同变异类型的稻瘟病菌菌株进行致病性测定。根据稻瘟病菌两种交配型MAT1-1和MAT1-2型分子标记,检测侵染美香占2号的稻瘟病菌可能存在的交配型。【结果】致病性分析结果显示,2013—2018年采集自美香占2号品种的52个稻瘟病菌菌株对其中的4个单基因系IRBL9-W(Pi9)、IRBLzt-T(Piz-t)、NIL-e1(Pi50)和IRBLkh-K3(Pikh)均表现无毒;然而,侵染单基因系IRBLz-Fu(Piz)、IRBLkp-K60(Pikp)和IRBLi-F5(Pii)的有毒菌株频率维持在57%以上,且侵染单基因系IRBLz-Fu(Piz)的有毒菌株频率在不同年份间呈现显著性差异,表明美香占2号种植区田间稻瘟病菌群体中存在较高频率的avrPiz、avrPikp和avrPii毒性菌株。根据8个已克隆无毒基因的功能性分子标记检测,在52个供试菌株中均可检测到无毒基因AvrPiz-t、AvrPi9和AvrPik-CDE,但扩增不到Avr1-CO39、AvrPia和AvrPii,仅有4个菌株可检测到无毒基因AvrPita,有3个菌株可检测到无毒基因AvrPik-ABF。所测序7个菌株的AvrPi9编码区序列和AvrPiz-t包含启动子及编码区的序列与其各自无毒基因的序列完全一致;所测序7个菌株的AvrPik-CDE包含启动子及编码区的序列中,有4个菌株的序列与AvrPik无毒基因序列完全一致,有3个菌株在编码区存在CTTT连续4个碱基的插入。在来自广东不同稻区美香占2号的分离菌株群体中,共发现了4种不同的AvrPib 无毒基因扩增模式(扩增预期大小的条带、有转座子Pot3插入的条带、有转座子Mg-SINE插入的条带和无扩增条带),共呈现5种不同的单倍型(AvrPib-AP1-1、AvrPib-AP1-2、avrPib-AP2、avrPib-AP3和avrPib-AP2+avrPib-AP3),AvrPib-AP1-1和AvrPib-AP1-2是无毒型,而avrPib-AP2和avrPib-AP3则在基因的启动子区域分别存在Pot3或Mg-SINE的插入而呈现毒性型,在1个菌株中检测到avrPib-AP2+avrPib-AP3双单倍型。交配型分子标记分析结果表明,检测的52个稻瘟病菌菌株均为MAT1-2交配型,其中有2个菌株GD18-009和GD18-185采集于广东韶关稻区,同时可检测到MAT1-1交配型,具有双交配型。【结论】在广东美香占2号品种上分离的稻瘟病菌普遍含有无毒基因AvrPiz-t、AvrPi9、AvrPi50和AvrPikh,AvrPib的变异类型丰富多样,该研究结果可为优质品种美香占2号的科学布局及其种植区域其他不同抗病基因型品种的合理选择提供参考。
汪文娟,苏菁,陈深,杨健源,陈凯玲,冯爱卿,汪聪颖,封金奇,陈炳,朱小源. 广东省侵染美香占2号的稻瘟病菌致病性及无毒基因变异分析[J]. 中国农业科学, 2022, 55(7): 1346-1358.
WANG WenJuan,SU Jing,CHEN Shen,YANG JianYuan,CHEN KaiLing,FENG AiQing,WANG CongYing,FENG JinQi,CHEN Bing,ZHU XiaoYuan. Pathogenicity and Avirulence Genes Variation of Magnaporthe oryzae from a Rice Variety Meixiangzhan 2 in Guangdong Province[J]. Scientia Agricultura Sinica, 2022, 55(7): 1346-1358.
表1
采自广东的52 个稻瘟病菌菌株信息"
| 采集地 Sampling location | 菌株编号 Strain number | 采集地 Sampling location | 菌株编号 Strain number |
|---|---|---|---|
| 韶关Shaoguan | GD13-012 | 韶关Shaoguan | GD16-187 |
| 韶关Shaoguan | GD13-027 | 韶关Shaoguan | GD16-189 |
| 韶关Shaoguan | GD13-354 | 韶关Shaoguan | GD16-196 |
| 韶关Shaoguan | GD13-358 | 韶关Shaoguan | GD17-001 |
| 韶关Shaoguan | GD13-369 | 河源Heyuan | GD17-125 |
| 韶关Shaoguan | GD13-382 | 惠州Huizhou | GD17-165 |
| 韶关Shaoguan | GD13-389 | 惠州Huizhou | GD17-171 |
| 韶关Shaoguan | GD13-394 | 惠州Huizhou | GD17-181 |
| 韶关Shaoguan | GD13-395 | 韶关Shaoguan | GD17-312 |
| 河源Heyuan | GD13-452 | 韶关Shaoguan | GD17-313 |
| 阳江Yangjiang | GD13-591 | 韶关Shaoguan | GD17-314 |
| 河源Heyuan | GD14-073 | 惠州Huizhou | GD17-380 |
| 韶关Shaoguan | GD14-186 | 韶关Shaoguan | GD18-009 |
| 云浮Yunfu | GD14-295 | 惠州Huizhou | GD18-094 |
| 清远Qingyuan | GD15-074 | 韶关Shaoguan | GD18-185 |
| 清远Qingyuan | GD15-076 | 韶关Shaoguan | GD18-188 |
| 惠州Huizhou | GD15-097 | 韶关Shaoguan | GD18-189 |
| 韶关Shaoguan | GD15-195 | 韶关Shaoguan | GD18-191 |
| 韶关Shaoguan | GD15-196 | 韶关Shaoguan | GD18-192 |
| 韶关Shaoguan | GD15-198 | 韶关Shaoguan | GD18-193 |
| 韶关Shaoguan | GD15-205 | 韶关Shaoguan | GD18-194 |
| 韶关Shaoguan | GD15-206 | 韶关Shaoguan | GD18-196 |
| 韶关Shaoguan | GD15-214 | 惠州Huizhou | GD18-269 |
| 韶关Shaoguan | GD16-006 | 惠州Huizhou | GD18-271 |
| 河源Heyuan | GD16-034 | 惠州Huizhou | GD18-272 |
| 韶关Shaoguan | GD16-185 | 增城Zengcheng | GD18-361 |
表2
本研究中用于检测无毒基因和交配型的引物"
| 引物 Primer | 引物序列 Sequence (5′-3′) | 预期片段大小 Expected fragment size (bp) | 目的 Purpose |
|---|---|---|---|
| Avr1-CO39 F | TGCCGCATTTTGCTAACCG | 994 | 检测Avr1-CO39启动子及CDS To detect Avr1-CO39 promoter and CDS |
| Avr1-CO39 R | GCGAATCCATAGACAAGGAC | ||
| AvrPita F | CAGGCATACATTGGAGAGCC | 1549 | 检测AvrPita启动子及CDS To detect AvrPita promoter and CDS |
| AvrPita R | CCCTCCATTCCAACACTAAC | ||
| AvrPii F | GGTAGATATCCGCTGACTGG | 839 | 检测AvrPii启动子及CDS To detect AvrPii promoter and CDS |
| AvrPii R | ACTGTCCGCCGCTCGTTTGG | ||
| AvrPi9 F | ATGCAGTTCTCTCAGATCCTC | 342 | 检测AvrPi9 CDS To detect AvrPi9 CDS |
| AvrPi9 R | CTACCAGTGCGTCTTTTCGAC | ||
| MGG-AvrPiz-t F | AACCCGCAGCAGCATAAAAG | 1621 | 检测AvrPiz-t启动子及CDS To detect AvrPiz-t promoter and CDS |
| MGG-AvrPiz-t R | GAAAAGTGGCTCGTTCCTAATTG | ||
| AvrPia F | CAGAGAAACGGACTTGGAGG | 1220 | 检测AvrPia启动子及CDS To detect AvrPia promoter and CDS |
| AvrPia R | GGTATACACGTACGGTAGGG | ||
| AvrPik-CDE F | TCCTGCTGCTAACTCCATTC | ~1000 | 检测AvrPik-CDE型启动子及CDS To detect AvrPik-CDE promoter and CDS |
| AvrPik-CDE R | GGGTACAGGAATACCAGG | ||
| AvrPik-ABF F | TCCTGCTGCTAACTCCATTC | ~1000 | 检测AvrPik-ABF型启动子及CDS To detect AvrPik-ABF promoter and CDS |
| AvrPik-ABF R | GGGTACAGGAATATCAGC | ||
| MGG-AvrPib F2 | GGCCTATCGCATGTTTATATTGAG | 633 | 检测AvrPib启动子及部分CDS To detect AvrPib promoter and some CDS |
| MGG-AvrPib R2 | AGGCGATAGTGATAAAAGTGGTTG | ||
| MAT1-1F | GACATCAAGGCTCGTCGAC | 650 | 交配型分析 Mating type analysis |
| MAT1-1R | GAGTGCGCCATTAGGAAGC | ||
| MAT1-2F | GGACCTCGAATTCGTCGAC | 300 | 交配型分析 Mating type analysis |
| MAT1-2R | GGCATCTTTCAGGATAGACC |
表3
2013—2018年采集的稻瘟病菌群体中对抗病单基因系有毒菌株频率"
| 年份(菌株数) Year (Number of strains) | IRBL9-W | IRBLzt-T | NIL-e1 | IRBLkh-K3 | IRBL1-CL | IRBLta2-Re | IRBLz-Fu | IRBLkp-K60 | IRBLi-F5 | CK |
|---|---|---|---|---|---|---|---|---|---|---|
| Pi9 | Piz-t | Pi50 | Pikh | Pi1 | Pita2 | Piz | Pikp | Pii | LTH | |
| 2013 (11) | 0 | 0 | 0 | 0 | 0 | 9.09 | 81.82 | 81.82 | 72.73 | 100.00 |
| 2015 (9) | 0 | 0 | 0 | 0 | 0 | 33.33 | 88.89 | 88.89 | 77.78 | 100.00 |
| 2016 (6) | 0 | 0 | 0 | 0 | 16.67 | 16.67 | 100.00 | 66.67 | 83.33 | 100.00 |
| 2017 (9) | 0 | 0 | 0 | 0 | 33.33 | 22.22 | 77.78 | 88.89 | 77.78 | 100.00 |
| 2018 (14) | 0 | 0 | 0 | 0 | 14.29 | 21.43 | 57.14 | 78.57 | 78.57 | 100.00 |
| χ2 for homogeneity | 59.61*** | 15.25** | 12.37** | 4.16 | 0.73 |
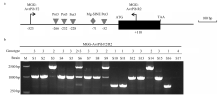
图2
分离自美香占2号稻瘟病菌中无毒基因AvrPib等位基因的变异 a:引物MGG-AvrPib F2/R2检测转座子Pot3和 Mg-SINE在AvrPib(KM887844.1)中的插入 The positions of the Pot3 and Mg-SINE transposon detected by the primer pair MGG-AvrPib F2/R2 within the AvrPib sequence (KM887844.1);b:17 个测序稻瘟病菌菌株的基因型,1—3(1条可见带型)、4(缺失)Genotyping of 17 sequenced M. oryzae strains. 1-3 (one fragment of variable size), 4 (absent) "
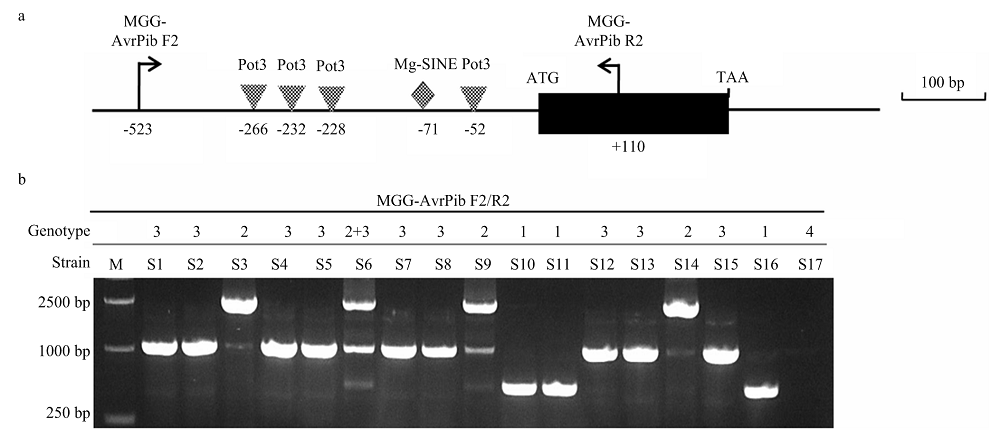
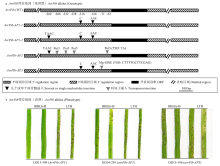
图3
分离自美香占2号稻瘟病菌无毒基因AvrPib等位基因序列变异特征 a:AvrPib等位基因变异特征Characterization of allelic variations at AvrPib。WT:野生型 Wild type (AvrPib-KM887844.1);AP1-1:TAACGT插入 TAACGT insertion;AP1-2:TAAC 插入 TAAC insertion;AP2:Pot3插入 Pot3 insertion;AP3:Mg-SINE 插入 Mg-SINE insertion。b:具有不同 AvrPib 等位基因变异型菌株的表型The phenotype of strains with different AvrPib allelic variations"

表4
15个代表菌株的AvrPib单倍型特征"
| AvrPib等位基因的核苷酸多态性Nucleotide polymorphisms of AvrPib allelesa | |||||||
|---|---|---|---|---|---|---|---|
| -269 bp | -266 bp/-232 bp/-228 bp/-52 bp | -216 bp | -124 bp | -71 bp | |||
| 单倍型 Haplotype | 频率 Frequency (%) | 表型 Pathotype to IRBLb-B (Pib) | - - - - | - | - - - | ||
| AvrPib-AP1-1 | 20.00 | A | - - - - | C | AAC | ||
| AvrPib-AP1-2 | 6.67 | A | TAAC | C | AAC | ||
| avrPib-AP2 | 33.33 | V | - - - -或TAAC | Pot3 | |||
| avrPib-AP3 | 33.33 | V | - - - - | C | AAC | Mg-SINE | |
| avrPib-AP2+avrPib-AP3 | 6.67 | V | - - - - | Pot3 | C | AAC | Mg-SINE |
| [1] | KHUSH G S, JENA K K. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.)//WANG G L, VALENT B. Advances in Genetics, Genomics and Control of Rice Blast Disease. New York: Springer, 2009: 1-10. |
| [2] |
ZHANG H F, ZHENG X B, ZHANG Z G. The Magnaporthe grisea species complex and plant pathogenesis. Molecular Plant Pathology, 2016, 17(6):796-804.
doi: 10.1111/mpp.12342 |
| [3] |
SAVARY S, WILLOCQUET L, PETHYBRIDGE S J, ESKER P, MCROBERTS N, NELSON A. The global burden of pathogens and pests on major food crops. Nature Ecology and Evolution, 2019, 3(3):430-439.
doi: 10.1038/s41559-018-0793-y |
| [4] | ASHKANI S, RAFII M Y, SHABANIMOFRAD M, MIAH G, SAHEBI M, AZIZI P, TANWEER F A, AKHTAR M S, NASEHI A. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Frontiers in Plant Science, 2015, 6:886. |
| [5] |
TANWEER F A, RAFII M Y, SIJAM K, RAHIM H A, AHMED F, LATIF M A. Current advance methods for the identification of blast resistance genes in rice. Comptes Rendus Biologies, 2015, 338(5):321-334.
doi: 10.1016/j.crvi.2015.03.001 |
| [6] |
KIRZINGER M W, STAVRINIDES J. Host specificity determinants as a genetic continuum. Trends in Microbiology, 2012, 20(2):88-93.
doi: 10.1016/j.tim.2011.11.006 |
| [7] | 赵雷, 周少川, 王重荣, 李宏, 黄道强, 王志东, 周德贵, 陈宜波, 吴玉坤. 绿色广适性优质稻品种的系谱分析及育种应用研究. 生命科学, 2018, 30(10):1100-1107. |
| ZHAO L, ZHOU S C, WANG C R, LI H, HUANG D Q, WANG Z D, ZHOU D G, CHEN Y B, WU Y K. Pedigree analysis and breeding and application of green widely adaptive good quality rice varieties. Chinese Bulletin of Life Sciences, 2018, 30(10):1100-1107. (in Chinese) | |
| [8] | 汪文娟, 苏菁, 杨健源, 陈深, 鲁国东, 朱小源. 侵染优质稻美香占2号的稻瘟病菌生理小种鉴定及无毒基因分析. 植物保护学报, 2020, 47(3):572-582. |
| WANG W J, SU J, YANG J Y, CHEN S, LU G D, ZHU X Y. Identification of physiological race and analysis avirulent genes for isolates of rice blast infecting from rice variety of Meixiangzhan 2. Journal of Plant Protection, 2020, 47(3):572-582. (in Chinese) | |
| [9] |
FLOR H H. Current status of the gene-for-gene concept. Annual Review of Phytopathology, 1971, 9:275-296.
doi: 10.1146/annurev.py.09.090171.001423 |
| [10] |
ORBACH M J, FARRALL L, SWEIGARD J A, CHUMLEY F G, VALENT B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. The Plant Cell, 2000, 12(11):2019-2032.
doi: 10.1105/tpc.12.11.2019 |
| [11] |
VALENT B, KHANG C H. Recent advances in rice blast effector research. Current Opinion in Plant Biology, 2010, 13(4):434-441.
doi: 10.1016/j.pbi.2010.04.012 |
| [12] | DODDS P N, LAWRENCE G J, CATANZARITI A M, TEH T, WANG C A, AYLIFFE M A, KOBE B, ELLIS J G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proceedings of National Academy of Sciences of the United States of America, 2006, 103(23):8888-8893. |
| [13] |
YOSHIDA K, SAITOH H, FUJISAWA S, KANZAKI H, MATSUMURA H, YOSHIDA K, TOSA Y, CHUMA I, TAKANO Y, WIN J, KAMOUN S, TERAUCHI R. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. The Plant Cell, 2009, 21(5):1573-1591.
doi: 10.1105/tpc.109.066324 |
| [14] |
KANZAKI H, YOSHIDA K, SAITOH H, FUJISAKI K, HIRABUCHI A, ALAUX L, FOURNIER E, THARREAU D, TERAUCHI R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. The Plant Journal, 2012, 72(6):894-907.
doi: 10.1111/j.1365-313X.2012.05110.x |
| [15] |
WU J, KOU Y J, BAO J D, LI Y, TANG M Z, ZHU X L, PONAYA A, XIAO G, LI J B, LI C Y, et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytologist, 2015, 206(4):1463-1475.
doi: 10.1111/nph.13310 |
| [16] |
ZHANG S L, WANG L, WU W H, HE L Y, YANG X F, PAN Q H. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Scientific Reports, 2015, 5:11642.
doi: 10.1038/srep11642 |
| [17] | RAY S, SINGH P K, GUPTA D K, MAHATO A K, SARKAR C, RATHOUR R, SINGH N K, SHARMA T R. Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Frontiers in Plant Science, 2016, 7:1140. |
| [18] |
WANG B H, EBBOLE D J, WANG Z H. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. Journal of Integrative Agriculture, 2017, 16(12):2746-2760.
doi: 10.1016/S2095-3119(17)61746-5 |
| [19] |
SELISANA S M, YANORIA M J, QUIME B, CHAIPANYA C, LU G, OPULENCIA R, WANG G L, MITCHELL T, CORRELL J, TALBOT N J, LEUNG H, ZHOU B. Avirulence (AVR) gene-based diagnosis complements existing pathogen surveillance tools for effective deployment of resistance (R) genes against rice blast disease. Phytopathology, 2017, 107(6):711-720.
doi: 10.1094/PHYTO-12-16-0451-R |
| [20] |
OLUKAYODE T, QUIME B, SHEN Y C, YANORIA M J, ZHANG S B, YANG J Y, ZHU X Y, SHEN W C, VON TIEDEMANN A, ZHOU B. Dynamic insertion of Pot3 in AvrPib prevailing in a field rice blast population in the Philippines lead to the high virulence frequency against the resistance gene Pib in rice. Phytopathology, 2019, 109(5):870-877.
doi: 10.1094/PHYTO-06-18-0198-R |
| [21] |
WANG W J, SU J, CHEN K L, YANG J Y, CHEN S, WANG C Y, FENG A Q, WANG Z H, WEI X Y, ZHU X Y, LU G D, ZHOU B. Dynamics of the rice blast fungal population in the field after deployment of an improved rice variety containing known resistance genes. Plant Disease, 2021, 105(4):919-928.
doi: 10.1094/PDIS-06-20-1348-RE |
| [22] |
LATORRE S M, REYES-AVILA C S, MALMGREN A, WIN J, KAMOUN S, BURBANO H A. Differential loss of effector genes in three recently expanded pandemic clonal lineages of the rice blast fungus. BMC Biology, 2020, 18(1):88.
doi: 10.1186/s12915-020-00818-z |
| [23] |
YOSHIDA K, SAUNDERS D G, MITSUOKA C, NATSUME S, KOSUGI S, SAITOH H, INOUE Y, CHUMA I, TOSA Y, CANO L M, KAMOUN S, TERAUCHI R. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics, 2016, 17:370.
doi: 10.1186/s12864-016-2690-6 |
| [24] |
XING J, JIA Y, CORRELL J C, LEE F N, CARTWRIGHT R, CAO M, YUAN L. Analysis of genetic and molecular identity among field isolates of the rice blast fungus with an international differential system, Rep-PCR, and DNA sequencing. Plant Disease, 2013, 97(4):491-495.
doi: 10.1094/PDIS-04-12-0344-RE |
| [25] |
CHEN Q H, WANG Y C, ZHENG X B. Genetic diversity of Magnaporthe grisea in China as revealed by DNA fingerprint haplotypes and pathotypes. Journal of Phytopathology, 2006, 154(6):361-369.
doi: 10.1111/j.1439-0434.2006.01106.x |
| [26] |
GEORGE M L, NELSON R J, ZEIGLER R S, LEUNG H. Rapid population analysis of Magnaporthe grisea by using rep-PCR and endogenous repetitive DNA sequences. Phytopathology, 1998, 88(3):223-229.
doi: 10.1094/PHYTO.1998.88.3.223 |
| [27] |
THUAN N T N, BIGIRIMANA J, ROUMEN E D, VAN DER STRAETEN D, HÖFTE M. Molecular and pathotype analysis of the rice blast fungus in North Vietnam. European Journal of Plant Pathology, 2006, 114(4):381-396.
doi: 10.1007/s10658-006-0002-8 |
| [28] |
IMAM J, ALAM S, MANDAL N P, SHUKLA P, SHARMA T R, VARIAR M. Molecular identification and virulence analysis of AVR genes in rice blast pathogen, Magnaporthe oryzae from Eastern India. Euphytica, 2015, 206:21-31.
doi: 10.1007/s10681-015-1465-5 |
| [29] |
LE M T, ARIE T, TERAOKA T. Population dynamics and pathogenic races of rice blast fungus, Magnaporthe oryzae in the Mekong Delta in Vietnam. Journal of General Plant Pathology, 2010, 76:177-182.
doi: 10.1007/s10327-010-0231-8 |
| [30] |
TSUNEMATSU H, YANORIA M J T, EBRON L A, HAYASHI N, ANDO I, KATO H, IMBE T, KHUSH G S. Development of monogenic lines of rice for rice blast resistance. Breeding Science, 2000, 50(3):229-234.
doi: 10.1270/jsbbs.50.229 |
| [31] |
WANG J C, CORRELL J C, JIA Y. Characterization of rice blast resistance genes in rice germplasm with monogenic lines and pathogenicity assays. Crop Protection, 2015, 72:132-138.
doi: 10.1016/j.cropro.2015.03.014 |
| [32] |
WANG J C, JIA Y, WEN J W, LIU W P, LIU X M, LI L, JIANG Z Y, ZHANG J H, GUO X L, REN J P. Identification of rice blast resistance genes using international monogenic differentials. Crop Protection, 2013, 45:109-116.
doi: 10.1016/j.cropro.2012.11.020 |
| [33] |
ZHANG Y L, WANG J Y, YAO Y X, JIN X H, CORRELL J, WANG L, PAN Q H. Dynamics of race structures of the rice blast pathogen population in Heilongjiang Province, China from 2006 through 2015. Plant Disease, 2019, 103(11):2759-2763.
doi: 10.1094/PDIS-10-18-1741-RE |
| [34] |
ZHU X Y, CHEN S, YANG J Y, ZHOU S C, ZENG L X, HAN J L, SU J, WANG L, PAN Q H. The identification of Pi50 (t), a new member of the rice blast resistance Pi2/Pi9 multigene family. Theoretical and Applied Genetics, 2012, 124(7):1295-1304.
doi: 10.1007/s00122-012-1787-9 |
| [35] | 马军韬, 张国民, 张丽艳, 邓凌韦, 王永力, 王英. 黑龙江省水稻种质抗瘟性及稻瘟病菌致病性分析. 植物保护学报, 2017, 44(2):209-216. |
| MA J T, ZHANG G M, ZHANG L Y, DENG L W, WANG Y L, WANG Y. Analysis of blast-resistance of rice germplasm and pathogenicity of rice blast fungus Magnaporthe oryzae in Heilongjiang Province. Journal of Plant Protection, 2017, 44(2):209-216. (in Chinese) | |
| [36] | 朱小源, 杨祁云, 杨健源, 雷财林, 王久林, 凌忠专. 抗稻瘟病单基因系对籼稻稻瘟病菌小种鉴别力分析. 植物病理学报, 2004, 34(4):361-368. |
| ZHU X Y, YANG Q Y, YANG J Y, LEI C L, WANG J L, LING Z Z. Differentiation ability of monogenic lines to Magnaporthe grisea in indica rice. Acta Phytopathologica Sinica, 2004, 34(4):361-368. (in Chinese) | |
| [37] |
WU W H, WANG L, ZHANG S, LIANG Y Q, ZHENG X L, YI K X, HE C P. Assessment of sensitivity and virulence fitness costs of the AvrPik alleles from Magnaporthe oryzae to isoprothiolane. Genetics and Molecular Research, 2014, 13(4):9701-9709.
doi: 10.4238/2014.November.24.1 |
| [38] |
LI J, WANG Q, LI C, BI Y, FU X, WANG R. Novel haplotypes and networks of AVR-Pik alleles in Magnaporthe oryzae. BMC Plant Biology, 2019, 19:204.
doi: 10.1186/s12870-019-1817-8 |
| [39] | 孟峰, 张亚玲, 靳学慧, 张晓玉, 姜军. 黑龙江省稻瘟病菌无毒基因AVR-Pib、AVR-Pik和AvrPiz-t的检测与分析. 中国农业科学, 2019, 52(23):4262-4273. |
| MENG F, ZHANG Y L, JIN X H, ZHANG X Y, JIANG J. Detection and analysis of Magnaporthe oryzae avirulence genes AVR-Pib, AVR-Pik and AvrPiz-t in Heilongjiang Province. Scientia Agricultura Sinica, 2019, 52(23):4262-4273. (in Chinese) | |
| [40] |
XIAO G, YANG J Y, ZHU X Y, WU J, ZHOU B. Prevalence of ineffective haplotypes at the rice blast resistance (R) gene loci in Chinese elite hybrid rice varieties revealed by sequence-based molecular diagnosis. Rice, 2020, 13(1):6.
doi: 10.1186/s12284-020-0367-x |
| [1] | 刘瑞, 赵羽涵, 付忠举, 顾欣怡, 王艳霞, 靳学慧, 杨莹, 吴伟怀, 张亚玲. 黑龙江省和海南省PWL基因家族在稻瘟病菌中的分布及变异[J]. 中国农业科学, 2023, 56(2): 264-274. |
| [2] | 吴云雨,肖宁,余玲,蔡跃,潘存红,李育红,张小祥,黄年生,季红娟,戴正元,李爱宏. 长江下游粳稻稻瘟病广谱抗性基因组合模式分析[J]. 中国农业科学, 2021, 54(9): 1881-1893. |
| [3] | 孙小芳,刘敏,潘婷敏,龚国淑. 四川玉米小斑病菌交配型组成与育性分析[J]. 中国农业科学, 2021, 54(12): 2547-2558. |
| [4] | 代玉立,甘林,滕振勇,杨静民,祁月月,石妞妞,陈福如,杨秀娟. 玉米大斑病菌和小斑病菌交配型多重PCR 检测方法的建立与应用[J]. 中国农业科学, 2020, 53(3): 527-538. |
| [5] | 孟峰,张亚玲,靳学慧,张晓玉,姜军. 黑龙江省稻瘟病菌无毒基因AVR-Pib、AVR-Pik和AvrPiz-t的检测与分析[J]. 中国农业科学, 2019, 52(23): 4262-4273. |
| [6] | 汪文娟,苏菁,杨健源,韦小燕,陈凯玲,陈珍,陈深,朱小源. 源于广8 A杂交稻组合的稻瘟病菌无毒基因型分析[J]. 中国农业科学, 2018, 51(24): 4633-4646. |
| [7] | 王加峰,刘浩,王慧,陈志强. 水稻NBS-LRR类抗稻瘟病蛋白Pik-h的互作蛋白筛选[J]. 中国农业科学, 2016, 49(3): 482-490. |
| [8] | 岳晓凤,阙亚伟,王政逸. 基于RNA-Seq的稻瘟病菌Δznf1突变体的表达谱分析[J]. 中国农业科学, 2016, 49(17): 3347-3358. |
| [9] | 王世维1, 2, 郑文静2, 赵家铭2, 魏松红1, 王妍1, 赵宝海1, 刘志恒1. 辽宁省稻瘟病菌无毒基因型鉴定及分析[J]. 中国农业科学, 2014, 47(3): 462-472. |
| [10] | 李洪浩, 彭化贤, 席亚东, 王晓黎, 刘波微. 四川马铃薯晚疫病菌交配型、生理小种、甲霜灵敏感性 及mtDNA单倍型组成分析[J]. 中国农业科学, 2013, 46(4): 728-736. |
| [11] | 郭丽媛1, 贾慧1, 曹志艳1, 谷守芹1, 孙淑琴2, 董金皋1. 玉米大斑病菌有性杂交后代的交配型与寄生适合度分化[J]. 中国农业科学, 2013, 46(19): 4058-4065. |
| [12] | 周 洁,郑祥梓,兰 斓,林成增,林雄杰,鲁国东,王宗华 . 稻瘟病菌假定的糖基水解酶62家族初步研究[J]. 中国农业科学, 2009, 42(8): 2754-2762 . |
| [13] | . 两套鉴别品种对云南稻瘟病菌株鉴别能力的比较[J]. 中国农业科学, 2009, 42(2): 486-491 . |
| [14] | 沈瑛1;李成云2. 稻瘟病菌遗传多样性的研究回顾与展望[J]. 中国农业科学, 2007, 40(增刊): 3100-3106. |
| [15] | 朱杰华,杨志辉,张凤国,杜洪忠,刘大群. 马铃薯晚疫病菌群体遗传结构研究进展[J]. 中国农业科学, 2007, 40(9): 1936-1942 . |
|
||