













中国农业科学 ›› 2020, Vol. 53 ›› Issue (1): 1-17.doi: 10.3864/j.issn.0578-1752.2020.01.001
郭书磊1,2,鲁晓民1,齐建双1,魏良明1,张新1,韩小花1,岳润清1,王振华1( ),铁双贵1(
),铁双贵1( ),陈彦惠2(
),陈彦惠2( )
)
收稿日期:2019-05-20
接受日期:2019-07-11
出版日期:2020-01-01
发布日期:2020-01-19
通讯作者:
王振华,铁双贵,陈彦惠
作者简介:郭书磊,E-mail:guosl1309@163.com。
基金资助:
ShuLei GUO1,2,XiaoMin LU1,JianShuang QI1,LiangMing WEI1,Xin ZHANG1,XiaoHua HAN1,RunQing YUE1,ZhenHua WANG1( ),ShuangGui TIE1(
),ShuangGui TIE1( ),YanHui CHEN2(
),YanHui CHEN2( )
)
Received:2019-05-20
Accepted:2019-07-11
Online:2020-01-01
Published:2020-01-19
Contact:
ZhenHua WANG,ShuangGui TIE,YanHui CHEN
摘要:
【目的】叶片宽度和长度等叶形特性是决定植株形态,进而影响种植密度的重要农艺性状,通过转录组测序技术筛选并挖掘玉米叶片形态建成相关的代谢路径及调控基因,为深入认识叶片发育的分子机理和鉴定叶宽、叶长候选基因奠定基础。【方法】以极端窄叶自交系NL409和宽叶自交系WB665为材料,利用RNA-Seq技术鉴定7叶期第七片叶近基部的差异表达基因(DEGs),通过生物信息学分析,筛选与叶片发育密切相关的代谢通路,利用qRT-PCR验证不同激素路径叶形相关基因的表达结果,并结合启动子区域的序列差异挖掘叶形功能基因。【结果】分析对照(WB665)和样品(NL409)高通量测序结果,在叶宽形成关键部位共筛选出5 199个DEGs,其中,2 264(43.55%)个基因表达上调,2 935(56.45%)个基因下调表达,下调基因明显多于上调基因;GO功能富集分析表明,差异基因主要富集在细胞膜相关的细胞组分中,涉及代谢过程和细胞响应刺激;KEGG富集分析表明,差异基因主要参与到核糖体、植物激素信号转导、苯丙烷类代谢、乙醛酸和二羧酸代谢等过程,其中核糖体、植物激素信号转导、鞘脂类代谢下调表达基因较多的路径与叶片发育密切相关。核糖体路径富集到多个PRS(PRESSED FLOWER)基因,分析发现PRS13(PFL2)可能在调控窄叶发育过程中发挥重要作用。鞘脂代谢路径富集的基因几乎全部下调表达,引起抑制叶片发育的AP1(APETALA1)类和MAPK(Mitogen-Activated Protein Kinase)类基因上调,以及促进叶片发育的LFY(LEAFY)下调,与窄叶发育受抑制的表型一致。植物激素信号转导路径富集到的油菜素内酯(BR)响应基因和赤霉素(GA)代谢基因下调,细胞分裂素(CTK)和大部分生长素(Auxin)响应基因上调,与窄叶中DELLA蛋白基因上调表达,抑制GA并促进CTK基因表达的作用模式一致。通过qRT-PCR对18个叶片发育相关基因进行分析,结果表明,其表达趋势与转录组结果一致,分析发现BR相关的ROT3、Auxin相关的NAL7-like、AGO7-like以及TCP类转录因子CYC/TB1等基因与窄叶的形成密切相关。【结论】明确了一些与玉米叶片发育密切相关的代谢路径,还发现植物激素间的动态平衡对叶片发育有着重要影响,尤其是生长素与油菜素内酯、细胞分裂素与赤霉素之间的相互作用对调控叶片形态可能发挥重要作用。
郭书磊,鲁晓民,齐建双,魏良明,张新,韩小花,岳润清,王振华,铁双贵,陈彦惠. 利用RNA-Seq发掘玉米叶片形态建成相关的调控基因[J]. 中国农业科学, 2020, 53(1): 1-17.
ShuLei GUO,XiaoMin LU,JianShuang QI,LiangMing WEI,Xin ZHANG,XiaoHua HAN,RunQing YUE,ZhenHua WANG,ShuangGui TIE,YanHui CHEN. Explore Regulatory Genes Related to Maize Leaf Morphogenesis Using RNA-Seq[J]. Scientia Agricultura Sinica, 2020, 53(1): 1-17.
表2
样品reads分布情况"
| 样品1) Sample | 原始序列 Raw reads | 过滤后序列 Clean reads | 高质量序列 Clean reads (%) | 比对到基因组序列 Mapped reads | 锚定比例 Mapped (%) | 单一位置序列 Unique mapped reads (%) | 过滤后Q30比例 Q30 after filter (%) |
|---|---|---|---|---|---|---|---|
| NL409-71 | 48570184 | 47805714 | 98.43 | 38240306 | 79.99 | 77.12 | 93.19 |
| NL409-72 | 45215054 | 44236906 | 97.84 | 34918002 | 78.93 | 76.23 | 92.06 |
| NL409-73 | 46822340 | 46156716 | 98.58 | 37211749 | 80.62 | 77.79 | 95.02 |
| WB665-71 | 45625722 | 44792028 | 98.17 | 34166045 | 77.96 | 73.34 | 92.53 |
| WB665-72 | 43848894 | 43120278 | 98.34 | 33618206 | 78.80 | 75.21 | 94.68 |
| WB665-73 | 47838874 | 47202952 | 98.67 | 37193868 | 78.02 | 76.00 | 95.38 |
表3
差异表达基因显著富集的代谢通路"
| 代谢通路 Pathway | 代谢通路ID Pathway ID | 差异基因数目 Counts of DEGs | P值 P-value |
|---|---|---|---|
| 油菜素内酯生物合成Brassinosteroid biosynthesis | map00905 | 7 | 0.00088 |
| 乙醛酸和二羧酸代谢Glyoxylate and dicarboxylate metabolism | map00630 | 25 | 0.00108 |
| 核糖体Ribosome | map03010 | 83 | 0.00464 |
| 苯丙烷类代谢Phenylpropanoid biosynthesis | map00940 | 33 | 0.00854 |
| 植物激素信号转导Plant hormone signal transduction | map04075 | 57 | 0.01099 |
| 苯并恶嗪类生物合成Benzoxazinoid biosynthesis | map00402 | 7 | 0.02726 |
| 谷胱甘肽代谢Glutathione metabolism | map00480 | 23 | 0.02847 |
| 鞘脂类代谢Sphingolipid metabolism | map00600 | 13 | 0.03916 |
| 二萜类生物合成Diterpenoid biosynthesis | map00904 | 6 | 0.04792 |
| 类黄酮生物合成Flavonoid biosynthesis | map00941 | 8 | 0.04716 |
| 苯乙烯类、二芳庚酸类和姜酚类生物合成Stilbenoid, diarylheptanoid and gingerol biosynthesis | map00945 | 7 | 0.04825 |
表4
鞘脂类代谢路径及其相关基因"
| 路径 pathway | 基因编号 Gene ID | 差异倍数 FC | 基因描述/注释 Gene description/annotation | |
|---|---|---|---|---|
| 鞘脂类代谢 Sphingolipid metabolism | GRMZM2G165613 | 0.00 | 下调Down | 丝氨酸软脂酰转移酶,长链碱基生物合成蛋白1b Serine palmitoyltransferase, Long chain base biosynthesis protein 1b |
| GRMZM2G444378 | 0.00 | 下调Down | 丝氨酸软脂酰转移酶,PKC激活蛋白磷酸酶1型抑制剂 Serine palmitoyltransferase, PKC-activated protein phosphatase-1 inhibitor | |
| GRMZM2G449763 | 0.00 | 下调Down | 丝氨酸软脂酰转移酶,胺核黄素氧化还原酶 Serine palmitoyltransferase, Flavin containing amine oxidoreductase | |
| GRMZM2G076398 | 0.00 | 下调Down | 丝氨酸软脂酰转移酶,长链碱基生物合成蛋白1b Serine palmitoyltransferase, Long chain base biosynthesis protein 1b | |
| GRMZM2G134248 | 0.24 | 下调Down | 丝氨酸软脂酰转移酶,长链碱基生物合成蛋白1b Serine palmitoyltransferase, Long chain base biosynthesis protein 1b | |
| GRMZM2G144985 | 0.33 | 下调Down | 丝氨酸软脂酰转移酶,长链碱基生物合成蛋白1b Serine palmitoyltransferase, Long chain base biosynthesis protein 1b | |
| GRMZM2G152888 | 0.30 | 下调Down | 丝氨酸软脂酰转移酶,长链碱基生物合成蛋白2a Serine palmitoyltransferase, Long chain base biosynthesis protein 2a | |
| GRMZM2G357734 | 0.02 | 下调Down | 丝氨酸软脂酰转移酶,胺核黄素氧化还原酶 Serine palmitoyltransferase, Flavin containing amine oxidoreductase | |
| GRMZM2G402368 | 0.01 | 下调Down | SUR2,鞘氨醇C4型单加氧酶2 SUR2, sphinganine C4-monooxygenase 2 | |
| GRMZM2G030118 | 0.11 | 下调Down | SUR2,鞘氨醇C4型单加氧酶2 SUR2, sphinganine C4-monooxygenase 2 | |
| GRMZM2G346455 | 0.46 | 下调Down | α-半乳糖苷酶,参与植物细胞壁 Alpha-galactosidase 1, involved in plant-type cell wall | |
| GRMZM2G095126 | 2.66 | 上调Up | α-半乳糖苷酶,参与植物细胞壁 Alpha-galactosidase 1, involved in plant-type cell wall | |
| GRMZM2G105922 | 2.27 | 上调Up | GBA2,非溶酶体葡糖神经酰胺酶 GBA2, non-lysosomal glucosylceramidase | |
| MAP激酶 MAP kinase | GRMZM2G135904 | 13.57 | 上调Up | MAPK12,丝裂原激活蛋白激酶家族蛋白 MAPK12, MAP kinase family protein |
| GRMZM2G459824 | 8.117 | 上调Up | MAPKKK2,丝裂原激活蛋白激酶激酶激酶ANP1 MAPKKK2, MAP kinase kinase kinase ANP1-like | |
| GRMZM5G878379 | 3.00 | 上调Up | MAPKK4,丝裂原激活蛋白激酶家族蛋白 MAPKK4, MAP kinase family protein | |
| GRMZM2G064601 | 2.49 | 上调Up | MAPKKK,丝裂原激活蛋白激酶激酶激酶YODA MAPKKK, MAP kinase kinase kinase YODA | |
| GRMZM2G062761 | 2.04 | 上调Up | MAPK17,丝裂原激活蛋白激酶家族蛋白 MAPK17, MAP kinase family protein | |
| AP1类 AP1-like | GRMZM2G055782 | 17.58 | 上调Up | MADS-box型转录因子27 MADS-box transcription factor 27-like |
| GRMZM2G147716 | 3.78 | 上调Up | AGL8/AP1,K-box和MADS-box型转录因子家族 AGL8/AP1, K-box region and MADS-box transcription factor family | |
| GRMZM2G070034 | 2.88 | 上调Up | MADS-box型转录因子56 MADS-box transcription factor 56-like | |
| LFY 类LFY-like | GRMZM2G162814 | 0.25 | 下调Down | 多叶,花分生组织蛋白,影响花和叶片的发育 Leafy, floral meristem identity protein, affect flower and leaf development |
表5
植物激素信号转导路径相关基因"
| 激素 Hormones | 基因编号 Gene ID | 差异倍数 FC(T7/C7) | 基因描述/注释 Gene description/annotation | |
|---|---|---|---|---|
| 生长素信号 Auxin signaling | GRMZM2G030710 | 3.38 | 上调Up | ARF,类生长素响应因子15,调控转录 ARF, Auxin response factor 15-like, regulation of transcription |
| GRMZM2G078274 | 2.54 | 上调Up | ARF,类生长素响应因子3,调控转录 ARF, Auxin response factor 3-like, regulation of transcription | |
| GRMZM5G874163 | 2.07 | 上调Up | ARF,类生长素响应因子3,激活生长素信号通路 ARF, Auxin response factor 3-like, auxin-activated signaling pathway | |
| GRMZM2G361376 | 0.00 | 下调Down | ARF,类生长素响应因子15,细胞响应生长素刺激 ARF, Auxin response factor 15-like, cellular response to auxin stimulus | |
| GRMZM5G853479 | 70.68 | 上调Up | 生长素响应蛋白IAA4,细胞响应生长素刺激 Auxin-responsive protein IAA4, cellular response to auxin stimulus | |
| GRMZM2G104176 | 7.53 | 上调Up | 生长素响应蛋白IAA2,激活生长素信号通路 Auxin-responsive protein IAA2, auxin-activated signaling pathway | |
| GRMZM2G077356 | 4.12 | 上调Up | 生长素响应蛋白IAA13,激活生长素信号通路 Auxin-responsive protein IAA13, auxin-activated signaling pathwa | |
| GRMZM2G079957 | 3.86 | 上调Up | 生长素响应蛋白IAA31,激活生长素信号通路 Auxin-responsive protein IAA31, auxin-activated signaling pathway | |
| GRMZM2G074427 | 0.12 | 下调Down | 生长素响应蛋白IAA18,细胞响应生长素刺激 Auxin-responsive protein IAA18, cellular response to auxin stimulus | |
| GRMZM2G079200 | 0.01 | 下调Down | 生长素响应蛋白IAA23,激活生长素信号通路 Auxin-responsive protein IAA23, auxin-activated signaling pathway | |
| GRMZM2G059544 | 0.02 | 下调Down | 生长素响应蛋白IAA25,细胞响应生长素刺激 Auxin-responsive protein IAA25, cellular response to auxin stimulus | |
| GRMZM2G429254 | 4.12 | 上调Up | SAUR家族生长素响应蛋白SAUR32,生长素代谢 SAUR family auxin-responsive protein SAUR32, auxin metabolism | |
| GRMZM2G391596 | 3.46 | 上调Up | SAUR家族生长素响应蛋白SAUR36,生长素代谢 SAUR family auxin-responsive protein SAUR36, auxin metabolism | |
| GRMZM2G361993 | 0.34 | 下调Down | SAUR家族生长素响应蛋白SAUR32,生长素代谢 SAUR family auxin-responsive protein SAUR32, auxin metabolism | |
| GRMZM2G475683 | 0.24 | 下调Down | SAUR家族生长素响应蛋白SAUR12,生长素代谢 SAUR family auxin-responsive protein SAUR12, auxin metabolism | |
| GRMZM2G410567 | 2.35 | 上调Up | 生长素响应GH3基因家族,响应生长素刺激 Auxin-responsive GH3 gene family, response to auxin stimulus | |
| GRMZM2G067022 | 4.17 | 上调Up | AUX1,类生长素转运蛋白1,跨膜转运体 AUX1, Auxin transporter-like protein 1, transmembrane transporter | |
| GRMZM2G127949 | 2.00 | 上调Up | AUX1,类生长素转运蛋白1,跨膜转运体 AUX1, Auxin transporter-like protein 1, transmembrane transporter | |
| 油菜素内酯信号BR signaling | GRMZM2G158775 | 0.39 | 下调Down | BRI1,油菜素内酯不敏感1,膜受体,激活BZR1 BRI1, brassinosteroid insensitive 1, membrane receptor, activation of BZR1 |
| AC194970.5_FG002 | 0.27 | 下调Down | BZR1,芸苔素唑拮抗1,油菜素内酯信号转导 BZR1, brassinazole-resistant 1, brassinosteroid signal transduction | |
| GRMZM2G127050 | 0.49 | 下调Down | BSK,油菜素内酯信号激酶,膜受体,BRI1激酶底物 BSK, BR-signaling kinase, membrane receptor, substrates of BRI1 kinase | |
| GRMZM2G164224 | 0.31 | 下调Down | BSK,油菜素内酯信号激酶,膜受体,BRI1激酶底物 BSK, BR-signaling kinase, membrane receptor, substrates of BRI1 kinase | |
| GRMZM2G054634 | 0.17 | 下调Down | BSK,油菜素内酯信号激酶,膜受体,BRI1激酶底物 BSK, BR-signaling kinase, membrane receptor, substrates of BRI1 kinase | |
| AC210669.3_FG001 | 0.05 | 下调Down | 木葡聚糖转移酶TCH4,细胞伸长和扩展 Xyloglucosyl transferase TCH4, cell elongation and expansion | |
| 油菜素内酯合成BR biosynthesis | GRMZM2G103773 | 11.89 | 上调Up | 油菜素内酯-6-氧化酶2,细胞膜固有成分 Brassinosteroid-6-oxidase 2, intrinsic component of membrane |
| GRMZM2G107199 | 5.83 | 上调Up | D11,细胞色素P450,细胞膜固有成分,氧化还原活性 D11, cytochrome P450, integral component of membrane, oxidoreductase activity | |
| GRMZM2G143235 | 5.57 | 上调Up | D2,类固醇3-氧化酶,细胞膜部分,氧化还原活性 D2, steroid 3-oxidase, membrane part, oxidoreductase activity | |
| GRMZM2G032896 | 2.78 | 上调Up | D2,类固醇3-氧化酶,细胞膜固有成分,氧化还原活性 D2, steroid 3-oxidase, intrinsic component of membrane, oxidoreductase activity | |
| GRMZM2G065635 | 0.35 | 下调Down | DWF4,类固醇22-α-羟化酶,细胞膜固有成分 DWF4, steroid 22-alpha-hydroxylase, intrinsic component of membrane | |
| GRMZM2G162737 | 0.04 | 下调Down | CPD,细胞色素P450,细胞膜固有成分,氧化还原活性 CPD, cytochrome P450, integral component of membrane, oxidoreductase activity | |
| GRMZM5G872368 | 0.03 | 下调Down | BR6OX2,油菜素内酯-6-氧化酶2,细胞膜固有成分 BR6OX2, brassinosteroid-6-oxidase 2, intrinsic component of membrane | |
| 细胞分裂素信号CTK signaling | GRMZM5G884914 | 73.11 | 上调Up | 双组分响应调节器ARR-B家族,响应刺激 Two-component response regulator ARR-B family, response to stimulus |
| GRMZM2G409974 | 2.32 | 上调Up | 双组分响应调节器ARR-B家族,响应刺激 Two-component response regulator ARR-B family, response to stimulus | |
| GRMZM2G013612 | 2.11 | 上调Up | 类双组分响应调节器ARR-10,调控转录 Two-component response regulator ARR10-like, regulation of transcription | |
| 赤霉素信号GAs | GRMZM2G013016 | 6.80 | 上调Up | GAI1型DELLA蛋白,膜上细胞器,信号转导 DELLA protein GAI1-like, membrane-bounded organelle, signal transduction |
| 赤霉素代谢 相关基因 GAmrg | GRMZM2G073779 | 0.38 | 下调Down | GRAS家族参与赤霉素代谢,调控植株生长与发育 GRAS family, as a player in GA metabolism, regulate plant growth and development |
| GRMZM2G073805 | 0.29 | 下调Down | GRAS家族参与赤霉素代谢,调控植株生长与发育 GRAS family, as a player in GA metabolism, regulate plant growth and development | |
| GRMZM2G098517 | 0.23 | 下调Down | GRAS家族参与赤霉素代谢,调控植株生长与发育 GRAS family, as a player in GA metabolism, regulate plant growth and development | |
| GRMZM2G018254 | 0.04 | 下调Down | GRAS家族参与赤霉素代谢,调控植株生长与发育 GRAS family, as a player in GA metabolism, regulate plant growth and development | |
| GRMZM2G070371 | 0.00 | 下调Down | GRAS家族参与赤霉素代谢,调控植株生长与发育 GRAS family, as a player in GA metabolism, regulate plant growth and development | |
| GRMZM2G110579 | 3.02 | 上调Up | GRAS家族参与赤霉素代谢,调控植株生长与发育 GRAS family, as a player in GA metabolism, regulate plant growth and development | |
| GRMZM2G036340 | 0.42 | 下调Down | GA 3氧化酶1,赤霉素3-β-双加氧酶活性 GA 3-oxidase 1, gibberellin 3-beta-dioxygenase activity | |
| GRMZM2G006964 | 0.3 | 下调Down | GA 2氧化酶6,赤霉素2-β-双加氧酶活性 GA 2-oxidase 6, gibberellin 2-beta-dioxygenase activity | |
| GRMZM2G323504 | 0.37 | 下调Down | GA受体GID1L2,羟基亚甲丁酸吡喃葡糖转化酶 GA receptor GID1L2, tuliposide A-converting enzyme | |
| GRMZM2G049675 | 0.29 | 下调Down | GA受体GID1L2,羟基亚甲丁酸吡喃葡糖转化酶 GA receptor GID1L2, tuliposide A-converting enzyme | |
| GRMZM2G156310 | 0.09 | 下调Down | GA受体GID1L2,羟基亚甲丁酸吡喃葡糖转化酶 GA receptor GID1L2, tuliposide A-converting enzyme | |
| AC203966.5_FG005 | 0.21 | 下调Down | GA 20氧化酶1,调控叶形和矮化 GA 20 oxidase 1, regulate leaves shape and dwarfism | |
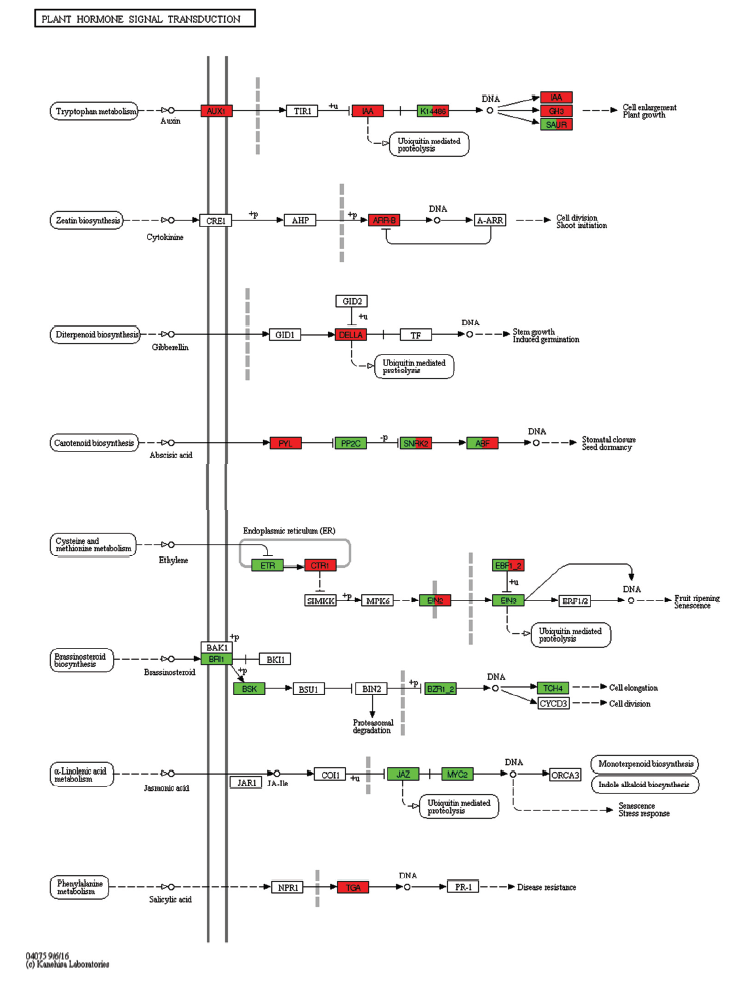
图6
植物激素信号转导 Tryptophan metabolism:色氨酸代谢;Auxin:生长素;Ubiquitin mediated proteolysis:泛素介导的蛋白水解;Cell enlargement:细胞增大;Plant growth:植物生长;Zeatin biosynthesis: 玉米素的生物合成;Cytokinine:细胞分裂素;Cell division:细胞分裂;Shoot initiation:根的起始;Diterpenoid biosynthesis:二萜类化合物的生物合成;Gibberellin:赤霉素;Stem growth:茎的生长;Induced germination:诱导萌发;Carotenoid biosynthesis:类胡萝卜素的生物合成;Abscisic acid:脱落酸;Stomatal closure:气孔闭合;Seed dormancy:种子休眠;Cysteine and methionine metabolism:半胱氨酸和蛋氨酸代谢;Ethylene:乙烯;Endoplasmic reticulum(ER):内质网;Fruit ripening:果实成熟;Senescence:衰老;Brassinosteroid biosynthesis:油菜素内酯生物合成;Brassinosteroid:油菜素甾醇;Proteasomal degradation:蛋白酶体降解;Cell elongation:细胞伸长;Alpha-Linolenic acid metabolism:α-亚麻酸代谢; Jasmonic acid:茉莉酸;Monoterpenoid biosynthesis类单萜生物合成;Indole alkaloid biosynthesis:吲哚生物碱生物合成;Stress response:应激反应;Phenylalanine metabolism:苯丙氨酸代谢;Salicylic acid:水杨酸;Disease resistance:抗病性。红色表示富集到的差异基因表达上调,绿色表示基因下调Red indicates that DEGs are up-regulated, and green indicates down-regulation of DEGs"
| [1] | 明博, 谢瑞芝, 侯鹏, 李璐璐, 王克如, 李少昆 . 2005—2016年中国玉米种植密度变化分析. 中国农业科学, 2017,50(11):1960-1972. |
| MING B, XIE R Z, HOU P, LI L L, WANG K R, LI S K . Changes of maize planting density in China. Scientia Agricultura Sinica, 2017,50(11):1960-1972. (in Chinese) | |
| [2] | 郭书磊, 陈娜娜, 齐建双, 岳润清, 韩小花, 燕树锋, 卢彩霞, 傅晓雷, 郭新海, 铁双贵 . 不同密度下玉米倒伏相关性状与产量的研究. 玉米科学, 2018,26(5):71-77. |
| GUO S L, CHEN N N, QI J S, YUE R Q, HAN X H, YAN S F, LU C X, FU X L, GUO X H, TIE S G . Study on the relationship between yield and lodging traits of maize under different planting densities. Journal of Maize Sciences, 2018,26(5):71-77. (in Chinese) | |
| [3] | YANG C W, XIE F M, JIANG Y P, LI Z, HUANG X, LI L . Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Developmental Cell, 2018,44(1):29-41. |
| [4] | 郭书磊, 张君, 齐建双, 岳润清, 韩小花, 燕树锋, 卢彩霞, 傅晓雷, 陈娜娜, 库丽霞, 铁双贵 . 玉米叶形相关性状的Meta-QTL及候选基因分析. 植物学报, 2018,53(4):487-501. |
| GUO S L, ZHANG J, QI J S, YUE R Q, HAN X H, YAN S F, LU C X, FU X L, CHEN N N, KU L X, TIE S G . Analysis of meta-quantitative trait loci and their candidate genes related to leaf shape in maize. Chinese Bulletin of Botany, 2018,53(4):487-501. (in Chinese) | |
| [5] | 崔晓峰, 黄海 . 叶发育的遗传调控机理研究进展. 植物生理学报, 2011,47(7):631-640. |
| CUI X F, HUANG H . Recent progresses from studies of leaf development. Plant Physiology Journal, 2011,47(7):631-640. (in Chinese) | |
| [6] | MORENO M A, HARPER L C, KRUEGER R W, DELLAPORTA S L , FREELING M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes & Development, 1997,11(5):616-628. |
| [7] | WALSH J, WATERS C A, FREELING M . The maize geneliguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes & Development, 1998,12(2):208-218. |
| [8] | ZHANG X L, MADI S, BORSUK L, NETTLETON D, ELSHIRE R J, BUCKNER B, BUCKNER D J, BECK J, TIMMERMANS M, SCHNABLE P S, SCANLON M J . Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genetics, 2007,3(6):1040-1052. |
| [9] | BOLDUC N, YILMAZ A, MEJIA-GUERRA M K, MOROHASHI K, O'CONNOR D, GROTEWOLD E, HAKE S, . Unraveling the KNOTTED1 regulatory network in maize meristems. Genes & Development, 2012,26(15):1685-1690. |
| [10] | TIMMERMANS M C, SCHULTES N P, JANKOVSKY J P, NELSON T . Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development, 1998,125(15):2813-2823. |
| [11] | DOUGLAS R N, WILEY D, SARKAR A, SPRINGER N, TIMMERMANS MARJA C P, SCANLON M J, . Ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. The Plant Cell, 2010,22(5):1441-1451. |
| [12] | CANDELA H, JOHNSTON R, GERHOLD A, FOSTER T, HAKE S . The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. The Plant Cell, 2008,20(8):2073-2087. |
| [13] | NELISSEN H, EECKHOUT D, DEMUYNCK K, PERSIAU G, WALTON A, BEL M V, VERVOORT M, CANDAELE J, BLOCK J D, AESAERT S, LIJSEBETTENS M V, GOORMACHTIG S, VANDEPOELE K, LEENE J V, MUSZYNSKI M, GEVAERT K, INZE D, JAEGER G D . Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. The Plant Cell, 2015,27(6):1605-1619. |
| [14] | WU L, ZHANG D, XUE M, QIAN J J, HE Y, WANG S C . Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. Journal of Integrative Plant Biology, 2014,56(11):1053-1063. |
| [15] | 吴庆飞, 秦磊, 董雷, 丁泽红, 李平华, 杜柏娟 . 玉米光合突变体hcf136(high chlorophyll fluorescence 136)的转录组分析. 作物学报, 2018,44(4):493-504. |
| WU Q F, QIN L, DONG L, DING Z H, LI P H, DU B J . Transcriptome analysis on a maize photosynthetic mutant hcf136 (high chlorophyll fluorescence 136). Acta Agronomica Sinica, 2018,44(4):493-504. (in Chinese) | |
| [16] | 邱化荣, 周茜茜, 何晓文, 张宗营, 张世忠, 陈学森, 吴树敬 . 基于转录组分析苹果水杨酸特异响应基因MdWRKY40的启动子鉴定. 中国农业科学, 2017,50(20):3970-3990. |
| QIU H R, ZHOU Q Q, HE X W, ZHANG Z Y, ZHANG S Z, CHEN X S, WU S J . Identification of MdWRKY40 promoter specific response to salicylic acid by transcriptome sequencing. Scientia Agricultura Sinica, 2017,50(20):3970-3990. (in Chinese) | |
| [17] | TRAPNELL C, WILLAMS B A, PERTEA G, MORTAZAVI A, KWAN G, BAREN M J, SALZBERG S L , WOLD B JHTER L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 2010,28(5):511-515. |
| [18] | WANG L K, FENG Z X, WANG X, WANG X W, ZHANG X G . DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics, 2009,26(1):136-138. |
| [19] | LIVAK K J, SCHMITTGEN T D . Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method . Methods, 2001,25(4):402-408. |
| [20] | SCHIPPERS J H M, MUELLER-ROEBER B . Ribosomal composition and control of leaf development. Plant Science, 2010,179(4):307-315. |
| [21] | YAO Y, LING Q H, WANG H, HUANG H . Ribosomal proteins promote leaf adaxial identity. Development, 2008,135(7):1325-1334. |
| [22] | SPIEGEL S, MERRILL Jr A H, . Sphingolipid metabolism and cell growth regulation. The FASEB Journal, 1996,10(12):1388-1397. |
| [23] | MERRILL Jr A H, SANDHOFF K . Metabolism and cell signaling//Biochemistry of Lipids, Lipoproteins and Mem-branes. Amsterdam, Elsevier Science Press, 2002, 373-407. |
| [24] | MONNIAUX M, MCKIM S M, CARTOLANO M, THEVENON E, PARCY F, MILTANTIS T, HAY A . Conservation vs divergence in LEAFY and APETALA1 functions between Arabidopsis thaliana and Cardamine hirsuta. New Phytologist, 2017,216(2):549-561. |
| [25] | LI P H, PONNALA L, GANDOTRA N, WANG L, SI Y Q, TAUSTA S L, KEBROM T H, PROVART N, PATEL R, MYERS C R, REIDEL E J, TURGEON R, LIU P, SUN Q, NELSON T, BRUTNELL T P . The developmental dynamics of the maize leaf transcriptome. Nature Genetics, 2010,42(12):1060-1067. |
| [26] | FACETTE M R, SHEN Z X, BJÖRNSDÓTTIR F R, BRIGGS S P, SMITH L G . Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. The Plant Cell, 2013: 2798-2812. |
| [27] | NICOLAS M, CUBAS P. The role of TCP transcription factors in shaping flower structure, leaf morphology, and plant architecture// Plant Transcription Factors: Evolutionary, Structural And Functional Aspects. Cambridge, MA: Academic Press, 2016: 249-267. |
| [28] | MANASSERO N G U, VIOLA I L, WELCHEN E, GONZALEZ D H . TCP transcription factors: architectures of plant form. Biomolecular Concepts, 2013,4(2):111-127. |
| [29] | KIM G T, TSUKAYA H, UCHIMIYA H . The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes & Development, 1998,12(15):2381-2391. |
| [30] | FUJIOKA S, LI J, CHOI Y H, SETO H, TAKATSUTO S, NOGUCHI T, WATANABE T, KURIYAMA H, YOKOT T, CHORY J, SAKURAI A . The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. The Plant Cell, 1997,9(11):1951-1962. |
| [31] | GUO Z, FUJIOKA S, BLANCAFLOR E B, MIAO S, GOU X P, LI J . TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. The Plant Cell, 2010,22(4):1161-1173. |
| [32] | SUZUK M, WANG H H Y, MCCARTY D R . Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiology, 2007,143(2):902-911. |
| [33] | HU Y X, WANG Y H, LIU X F, LI J Y . Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Research, 2004,14(1):8. |
| [34] | YUAN W Y, LUO X, LI Z C, YANG W N, WANG Y Z, LIU R, DU J M, HE Y H . A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nature Genetics, 2016,48(12):1527-1534. |
| [35] | JE B I, PIAO H L, PARK S J, PARK S H, KIM C M, XUAN Y H, PARK S H, HUANG J, CHOI Y D, AN G, WONG H L, FUJIOKA S, KIM M C, SHIMAMOTO K, HAN C . RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. The Plant Cell, 2010,22(6):1777-1791. |
| [36] | TIAN F, BRADBURY P J, BROWN P J, HUNG H Y, SUN Q, FLINT-GARCIN S, ROCHEFORD T R, MCMULLEN M D, HOLLAND J B, BUCKLER E S . Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature Genetics, 2011,43(2):159-165. |
| [37] | ISHIWATA A, OZAWA M, NAGASAKI H, KATO M, NODA Y, YAMAGUCHI T, NOSAKA M, SHIMIZU-SATO S, NAGASAKI A, MAEKAWA M, HIRANO H Y, SATO Y . Two WUSCHEL-related homeobox genes, narrow leaf2 and narrow leaf3, control leaf width in rice. Plant and Cell Physiology, 2013,54(5):779-792. |
| [38] | KUBO F C, YASUI Y, KUMAMARU T, SATO Y, HIRANO H Y . Genetic analysis of rice mutants responsible for narrow leaf phenotype and reduced vein number. Genes & Genetic Systems, 2016,91(4):235-240. |
| [39] | JIANG D, FANG J J, LOU L, ZHAO J F, YUAN S J, YIN L, SUN W, PENG L X, GUO B T, LI X Y . Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE, 2015,10(2):e0118169. |
| [40] | FUJINO K, MATSUDA Y, OZAWA K, NISHIMURA T, KOSHIBA T, FRAAIJE M W, SEKIGUCHI H . NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Molecular Genetics and Genomics, 2008,279(5):499-507. |
| [41] | DOUGLAS R N, WILEY D, SARKAR A, SPRINGER N, TIMMERMANS M C P, SCANLON M J, . Ragged seedling2 encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. The Plant Cell, 2010: 1441-1451. |
| [42] | DOTTO M C, PETSCH K A, AUKERMAN M J, BEATTY M, HAMMELL M, TIMMERMANS M C P . Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genetics, 2014,10(12):e1004826. |
| [43] | XIE Y, STRAUB D, EGUEN T, BRANDT R, STAHL M, Jaime F MARTINEZ-GAECIA J F WENKEL S . Meta-analysis of Arabidopsis KANADI1 direct target genes identifies a basic growth-promoting module acting upstream of hormonal signaling pathways. Plant Physiology, 2015,169(2):1240-1253. |
| [44] | EMERY J F, FLOYD S K, AIVAREZ J, ESHED Y, HAWKER N P, IZHAKI A, BAUM S F, BOWMAN J L . Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology, 2003,13(20):1768-1774. |
| [45] | OKUSHIMA Y, MITINA I, QUACH H L, THEOLOGIS A . AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. The Plant Journal, 2005,43(1):29-46. |
| [46] | SCHRUFF M C, SPIELMAN M, TIWARI S, ADAMS S, FENBY N, SCOTT R J . The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development, 2006,133(2):251-261. |
| [47] | TIAN H Y, LV B S, DING T T, BAI M Y, DING Z J . Auxin-BR interaction regulates plant growth and development. Frontiers in Plant Science, 2018,8:2256. |
| [48] | OH E, ZHU J Y, BAI M Y, ARENHART R A, SUN Y, WANG Z Y . Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife, 2014,3:e03031. |
| [49] | CHUNG Y, MAHARJAN P M, LEE O, FUJIOKA S, JANG S, KIM B, TAKATSUTO S, TSUJIMOTO M, KIM H, CHO S, PARK T, CHO H, HWANG I, CHOE S . Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. The Plant Journal, 2011,66(4):564-578. |
| [50] | SAKAMOTO T, MORINAKA Y, INUKAI Y, KITANO H, FUJIOKA S . Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. The Plant Journal, 2013,73(4):676-688. |
| [51] | JASINSKI S, PIAZZA P, CRAFT J, HAY A, WOOLLEY L, RIEU I, PHILLIPS A, HEDDEN P, TSIANTIS M . KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Current Biology, 2005,15(17):1560-1565. |
| [52] | NELISSEN H, RYMEN B, JIKUMARU Y, DEMUYNCK K, LIJSEBETTENS M V, KAMIYA Y, INZE D, BEEMSTER G T S . A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Current Biology, 2012,22(13):1183-1187. |
| [53] | FONOUNI-FARDE C, KISIALA A, BRAULT M, EMERY R J N, ANOUCK D, FLORIAN F . DELLA1-mediated gibberellin signaling regulates cytokinin-dependent symbiotic nodulation. Plant Physiology, 2017,175(4):1795-1806. |
| [54] | GREENBOIM-WAINBERG Y, MAYMON I, BOROCHOV R, ALVAREZ J, OLSZEWSKI N, ORI N, ESHED Y, WEISS D . Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. The Plant Cell, 2005,17(1):92-102. |
| [55] | WALSH J, WATERS C A, FREELING M . The maize geneliguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes & Development, 1998,12(2):208-218. |
| [56] | WANG Q L, XUE X J, LI Y L, DONG Y B, ZHANG L, ZHOU Q, DENG F, MA Z Y, QIAO D H, HU C H, REN Y L . A maize ADP-ribosylation factor ZmArf2 increases organ and seed size by promoting cell expansion in Arabidopsis. Physiologia Plantarum, 2016,156(1):97-107. |
| [1] | 柴海燕,贾娇,白雪,孟玲敏,张伟,金嵘,吴宏斌,苏前富. 吉林省玉米穗腐病致病镰孢菌的鉴定与部分菌株对杀菌剂的敏感性[J]. 中国农业科学, 2023, 56(1): 64-78. |
| [2] | 赵政鑫,王晓云,田雅洁,王锐,彭青,蔡焕杰. 未来气候条件下秸秆还田和氮肥种类对夏玉米产量及土壤氨挥发的影响[J]. 中国农业科学, 2023, 56(1): 104-117. |
| [3] | 李周帅,董远,李婷,冯志前,段迎新,杨明羡,徐淑兔,张兴华,薛吉全. 基于杂交种群体的玉米产量及其配合力的全基因组关联分析[J]. 中国农业科学, 2022, 55(9): 1695-1709. |
| [4] | 熊伟仡,徐开未,刘明鹏,肖华,裴丽珍,彭丹丹,陈远学. 不同氮用量对四川春玉米光合特性、氮利用效率及产量的影响[J]. 中国农业科学, 2022, 55(9): 1735-1748. |
| [5] | 李易玲,彭西红,陈平,杜青,任俊波,杨雪丽,雷鹿,雍太文,杨文钰. 减量施氮对套作玉米大豆叶片持绿、光合特性和系统产量的影响[J]. 中国农业科学, 2022, 55(9): 1749-1762. |
| [6] | 邱一蕾,吴帆,张莉,李红亮. 亚致死剂量吡虫啉对中华蜜蜂神经代谢基因表达的影响[J]. 中国农业科学, 2022, 55(8): 1685-1694. |
| [7] | 马小艳,杨瑜,黄冬琳,王朝辉,高亚军,李永刚,吕辉. 小麦化肥减施与不同轮作方式的周年养分平衡及经济效益分析[J]. 中国农业科学, 2022, 55(8): 1589-1603. |
| [8] | 李前,秦裕波,尹彩侠,孔丽丽,王蒙,侯云鹏,孙博,赵胤凯,徐晨,刘志全. 滴灌施肥模式对玉米产量、养分吸收及经济效益的影响[J]. 中国农业科学, 2022, 55(8): 1604-1616. |
| [9] | 张家桦,杨恒山,张玉芹,李从锋,张瑞富,邰继承,周阳晨. 不同滴灌模式对东北春播玉米籽粒淀粉积累及淀粉相关酶活性的影响[J]. 中国农业科学, 2022, 55(7): 1332-1345. |
| [10] | 谭先明,张佳伟,王仲林,谌俊旭,杨峰,杨文钰. 基于PLS的不同水氮条件下带状套作玉米产量预测[J]. 中国农业科学, 2022, 55(6): 1127-1138. |
| [11] | 冯宣军, 潘立腾, 熊浩, 汪青军, 李静威, 张雪梅, 胡尔良, 林海建, 郑洪建, 卢艳丽. 南方地区120份甜、糯玉米自交系重要目标性状和育种潜力分析[J]. 中国农业科学, 2022, 55(5): 856-873. |
| [12] | 刘苗,刘朋召,师祖姣,王小利,王瑞,李军. 氮磷配施下夏玉米临界氮浓度稀释曲线的构建与氮营养诊断[J]. 中国农业科学, 2022, 55(5): 932-947. |
| [13] | 乔远,杨欢,雒金麟,汪思娴,梁蓝月,陈新平,张务帅. 西北地区玉米生产投入及生态环境风险评价[J]. 中国农业科学, 2022, 55(5): 962-976. |
| [14] | 黄兆福, 李璐璐, 侯梁宇, 高尚, 明博, 谢瑞芝, 侯鹏, 王克如, 薛军, 李少昆. 不同种植区玉米生理成熟后田间站秆脱水的积温需求[J]. 中国农业科学, 2022, 55(4): 680-691. |
| [15] | 石习, 宁丽华, 葛敏, 邬奇, 赵涵. 玉米氮状况相关生物标记物的筛选和应用[J]. 中国农业科学, 2022, 55(3): 438-450. |
|
||