













中国农业科学 ›› 2022, Vol. 55 ›› Issue (19): 3751-3766.doi: 10.3864/j.issn.0578-1752.2022.19.006
收稿日期:2022-04-28
接受日期:2022-05-29
出版日期:2022-10-01
发布日期:2022-10-10
通讯作者:
吴志明
作者简介:康忱,E-mail: 基金资助:
KANG Chen( ),ZHAO XueFang,LI YaDong,TIAN ZheJuan,WANG Peng,WU ZhiMing(
),ZHAO XueFang,LI YaDong,TIAN ZheJuan,WANG Peng,WU ZhiMing( )
)
Received:2022-04-28
Accepted:2022-05-29
Online:2022-10-01
Published:2022-10-10
Contact:
ZhiMing WU
摘要:
【目的】对黄瓜全基因组的CC-NBS-LRR(CNL)基因家族进行生物信息学和表达模式分析,为深入研究CNL基因家族在黄瓜生长、发育和病害胁迫响应中的功能提供参考。【方法】以拟南芥CNL为参考序列,利用本地Perl语言和Pfam等软件检索黄瓜‘9930’基因组并确定黄瓜CNL基因家族成员。通过ExPASy、GSDS2.0、MEGA、MEME、Tbtools、Mev等工具对黄瓜CNL家族基因进行生物信息学分析。根据转录组数据库、霜霉病菌(Pseudoperonospora cubensis)、白粉病菌(Podosphaera xanthii)接种处理和实时荧光定量PCR技术分析该类基因的表达模式。【结果】从黄瓜全基因组中鉴定得到17个CsCNL,这些基因分布在除1号染色体外的6条染色体上,其编码蛋白与其他植物CNL结构相似,均含有CC、NBS和LRR保守结构域,蛋白质大小介于197—1 148 aa,分子量在22.6—131.3 kD,等电点在5.71—8.38。共线性分析表明其中不存在片段重复和串联重复基因,多物种系统进化关系表明,CsCNL基因家族成员在葫芦科植物之间具有较高的结构和功能相似性。CsCNL启动子中存在多种与抗病相关的顺式作用元件。CsCNL表达具有组织特异性。在接种霜霉病菌2 d和5 d后,Csa3G815400、Csa3G822360和Csa7G420890在抗性品种中均显著上调表达,Csa4G015850在抗性品种中下调表达,在敏感品种中显著性表现不一;在接种白粉病菌2 d和5 d后,Csa2G008000和Csa3G684170在敏感品种中显著上调表达,在抗性品种中显著下调表达;Csa4G016360、Csa7G420890和Csa7G425940在抗性品种中显著上调表达,在敏感品种中表达量无变化或显著下调。【结论】CsCNL家族成员具有组织表达特异性并且多数基因能够响应霜霉病菌和白粉病菌的胁迫。推测Csa3G815400、Csa3G822360和Csa7G420890表达量增加,Csa4G015850表达量减少可诱发抗病黄瓜品种的霜霉病抗病反应;Csa4G016360、Csa7G420890和Csa7G425940表达量增加,Csa2G008000和Csa3G684170表达量减少可诱发抗病黄瓜品种的白粉病抗病反应;而Csa7G420890表达量的增加能同时诱发抗病黄瓜品种的霜霉病和白粉病的抗病反应。
康忱,赵雪芳,李亚栋,田哲娟,王鹏,吴志明. 黄瓜CC-NBS-LRR家族基因鉴定及在霜霉病和白粉病胁迫下的表达分析[J]. 中国农业科学, 2022, 55(19): 3751-3766.
KANG Chen,ZHAO XueFang,LI YaDong,TIAN ZheJuan,WANG Peng,WU ZhiMing. Genome-Wide Identification and Analysis of CC-NBS-LRR Family in Response to Downy Mildew and Powdery Mildew in Cucumis sativus[J]. Scientia Agricultura Sinica, 2022, 55(19): 3751-3766.
表1
实时荧光定量PCR引物序列"
| 基因名称 Gene name | 正向引物序列 Forward sequence (5′-3′) | 反向引物序列 Reverse sequence (5′-3′) |
|---|---|---|
| ACTIN | TCCACGAGACTACCTACAACTC | GCTCATACGGTCAGCGAT |
| Csa2G008000 | GCCGCTCAATTTTCTGGAGA | GAGTAAACCCGTGTGCCATC |
| Csa2G074130 | GATGGATTGGCAACCTCACC | AGGCCAATCTTCTCGTCTGT |
| Csa2G433340 | TGTTCATGAGGCTGACGACT | TTGCGCAACTTCAGTCTCTG |
| Csa3G172400 | GCAACCCAACAAATCCGTCT | CTCGCCTAAGGTCCTCGTAG |
| Csa3G684170 | GCTGACAAAACACCTCCACA | ATCTGCTCCCTTTGCTTCCT |
| Csa3G815400 | GGCTTGAAAACTCGGCAGAA | ACTGCCAAATTCCGGTGTTC |
| Csa3G822360 | TGGCATGGGAGTTGGAGAAT | TATGCTCCACTGTTCGTCGA |
| Csa4G015840 | CCGAGATTGACGAGCATTCC | TGCAAGCTTGGAGGGTAGTT |
| Csa4G015850 | TGGGTGGAGAAGCTTGAACA | CCATGATACTCGCTGCACAA |
| Csa4G016360 | GTGAACGTTGCAGCTCAGAA | CCCATCTGCCCATCTCTTCA |
| Csa4G016430 | GGGGTCTACGAGAAATGGGT | TTGAATGAACCCTTGTGCCA |
| Csa4G016460 | TGGGTGCCGTAAAATGACAC | GAAGTTGCTCAGTCACGCTC |
| Csa5G266890 | ACCCAACTTCCCCAACAACT | TGGCCTCTCTCGAAGGTAAC |
| Csa6G375730 | AGGACCATGAACTCGACACA | GCCTTGTGCCATCCATTGTT |
| Csa7G420890 | CGAATTTTGGGGCAAGGACA | CTGTTGTCCTTGCTGCCATT |
| Csa7G425930 | TGTTCAACGTTGCTGCAAATG | CAATCTTTGACTGCGTGGCT |
| Csa7G425940 | TGAGTTCTTGCCAGTGGAGT | AAGTGTCCGGAGATTGCTGA |
表2
CsCNL家族基因全基因组鉴定与分子特征分析"
| 基因编号 Gene ID | 染色体位置 Genomic location | 氨基酸数量 Number of amino acid (aa) | 外显子 Exon | 分子量 MW (Da) | 等电点 PI |
|---|---|---|---|---|---|
| Csa2G008000 | Chr02:1351990-1356361 | 823 | 5 | 95328.28 | 6.80 |
| Csa2G074130 | Chr02:5810883-5814239 | 915 | 2 | 105955.85 | 8.29 |
| Csa2G433340 | Chr02:22624852-22628165 | 966 | 2 | 111929.62 | 8.38 |
| Csa3G172400 | Chr03:11532697-11536241 | 1148 | 1 | 131344.13 | 7.38 |
| Csa3G684170 | Chr03:26192533-26195851 | 1089 | 1 | 125618.99 | 6.10 |
| Csa3G815400 | Chr03:31465330-31468278 | 892 | 1 | 103116.33 | 6.43 |
| Csa3G822360 | Chr03:32245772-32249528 | 1053 | 2 | 120249.54 | 6.27 |
| Csa4G015840 | Chr04:2102886-2106281 | 1087 | 1 | 124356.65 | 6.84 |
| Csa4G015850 | Chr04:2107386-2110628 | 1080 | 1 | 123310.24 | 7.36 |
| Csa4G016360 | Chr04:2113346-2116883 | 958 | 2 | 109571.71 | 7.09 |
| Csa4G016430 | Chr04:2146414-2149099 | 840 | 3 | 96126.70 | 6.14 |
| Csa4G016460 | Chr04:2153828-2157080 | 947 | 2 | 108232.33 | 7.33 |
| Csa5G266890 | Chr05:11239765-11242908 | 1047 | 1 | 119379.10 | 6.04 |
| Csa6G375730 | Chr06:16934210-16937464 | 917 | 2 | 106784.55 | 8.21 |
| Csa7G420890 | Chr07:16247369-16250149 | 555 | 2 | 64716.85 | 7.61 |
| Csa7G425930 | Chr07:16263128-16264206 | 197 | 2 | 22566.76 | 5.71 |
| Csa7G425940 | Chr07:16268256-16272547 | 1020 | 5 | 116400.68 | 6.86 |
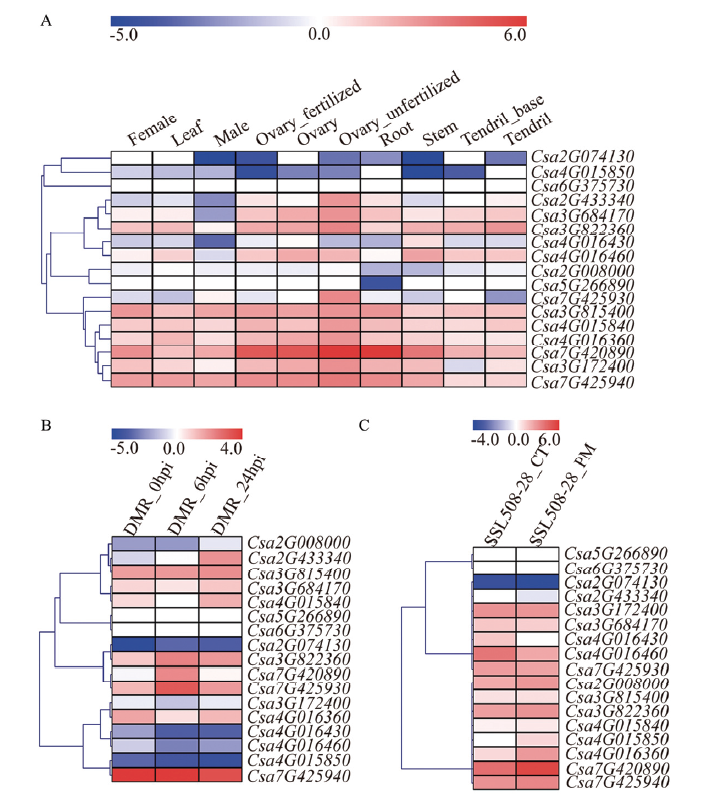
图7
CsCNL家族基因表达模式 A:组织表达模式Expression profiles of different tissues。Female:雌花Female flower;Leaf:叶;Male:雄花Male flower;Ovary_fertilized:受精子房;Ovary:子房;Ovary_unfertilized:未受精子房;Root:根;Stem:茎;Tendril_base:卷须基部;Tendril:卷须。B:在霜霉病菌处理下的表达模式Expression patterns after treated by P. cubensis。C:在白粉病菌处理下的表达模式Expression patterns after treated by P. xanthii"
| [1] |
BAKER C M, CHITRAKAR R, OBULAREDDY N, PANCHAL S, WILLIAMS P, MELOTTO M. Molecular battles between plant and pathogenic bacteria in the phyllosphere. Brazilian Journal of Medical and Biological Research, 2010, 43(8): 698-704.
doi: S0100-879X2010007500060 pmid: 20602017 |
| [2] |
HAAK D C, FUKAO T, GRENE R, HUA Z H, IVANOV R, PERRELLA G, LI S. Multilevel regulation of abiotic stress responses in plants. Frontiers in Plant Science, 2017, 8: 1564.
doi: 10.3389/fpls.2017.01564 pmid: 29033955 |
| [3] |
SONG W, FORDERER A, YU D L, CHAI J J. Structural biology of plant defence. New Phytologist, 2021, 229(2): 692-711.
doi: 10.1111/nph.16906 |
| [4] |
JONES J D G, DANGL J L. The plant immune system. Nature, 2006, 444(7117): 323-329.
doi: 10.1038/nature05286 |
| [5] |
BOLLER T, HE S Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 2009, 324(5928): 742-744.
doi: 10.1126/science.1171647 pmid: 19423812 |
| [6] |
TAKKEN F L W, TAMELING W I L. To nibble at plant resistance proteins. Science, 2009, 324(5928): 744-746.
doi: 10.1126/science.1171666 pmid: 19423813 |
| [7] |
WAN H J, YUAN W, YE Q J, WANG R Q, RUAN M Y, LI Z M, ZHOU G Z, YAO Z P, ZHAO J, LIU S J, YANG Y J. Analysis of TIR- and non-TIR-NBS-LRR disease resistance gene analogous in pepper: Characterization, genetic variation, functional divergence and expression patterns. BMC Genomics, 2012, 13: 502.
doi: 10.1186/1471-2164-13-502 pmid: 22998579 |
| [8] |
THOMAS A, CARBONE I, CHOE K, QUESADA-OCAMPO L M, OJIAMBO P S. Resurgence of cucurbit downy mildew in the United States: Insights from comparative genomic analysis of Pseudoperonospora cubensis. Ecology and Evolution, 2017, 7(16): 6231-6246.
doi: 10.1002/ece3.3194 |
| [9] |
PÉREZ-GARCÍA A, ROMERO D, FERNÁNDEZ-ORTUÑO D, LÓPEZ-RUIZ F, DE VICENTE A, TORÉS J A. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Molecular Plant Pathology, 2009, 10(2): 153-160.
doi: 10.1111/j.1364-3703.2008.00527.x |
| [10] |
BERG J A, APPIANO M, SANTILLÁN MARTÍNEZ M, HERMANS F W, VRIEZEN W H, VISSER R G, BAI Y L, SCHOUTEN H J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biology, 2015, 15: 243.
doi: 10.1186/s12870-015-0635-x |
| [11] |
SARASTE M, SIBBALD P R, WITTINGHOFER A. The P-loop—A common motif in ATP- and GTP-binding proteins. Trends in Biochemical Sciences, 1990, 15(11): 430-434.
doi: 10.1016/0968-0004(90)90281-F |
| [12] |
DANGL J L, JONES J D G. Plant pathogens and integrated defence responses to infection. Nature, 2001, 411(6839): 826-833.
doi: 10.1038/35081161 |
| [13] |
MATSUSHIMA N, MIYASHITA H. Leucine-rich repeat (LRR) domains containing intervening motifs in plants. Biomolecules, 2012, 2(2): 288-311.
doi: 10.3390/biom2020288 pmid: 24970139 |
| [14] |
SHAO Z Q, ZHANG Y M, HANG Y Y, XUE J Y, ZHOU G C, WU P, WU X Y, WU X Z, WANG Q, WANG B, CHEN J Q. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiology, 2014, 166(1): 217-234.
doi: 10.1104/pp.114.243626 |
| [15] |
SUKARTA O C A, SLOOTWEG E J, GOVERSE A. Structure- informed insights for NLR functioning in plant immunity. Seminars in Cell and Developmental Biology, 2016, 56: 134-149.
doi: 10.1016/j.semcdb.2016.05.012 |
| [16] |
ZHOU T, WANG Y, CHEN J Q, ARAKI H, JING Z, JIANG K, SHEN J, TIAN D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Molecular Genetics and Genomics, 2004, 271(4): 402-415.
doi: 10.1007/s00438-004-0990-z |
| [17] |
KANG Y J, KIM K H, SHIM S, YOON M Y, SUN S, KIM M Y, VAN K, LEE S. Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biology, 2012, 12: 139.
doi: 10.1186/1471-2229-12-139 pmid: 22877146 |
| [18] |
LOZANO R, PONCE O, RAMIREZ M, MOSTAJO N, ORJEDA G. Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS ONE, 2012, 7(4): e34775.
doi: 10.1371/journal.pone.0034775 |
| [19] |
LOZANO R, HAMBLIN M T, PROCHNIK S, JANNINK J. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics, 2015, 16: 360.
doi: 10.1186/s12864-015-1554-9 pmid: 25948536 |
| [20] | YANG X P, WANG J P. Genome-wide analysis of NBS-LRR genes in sorghum genome revealed several events contributing to NBS-LRR gene evolution in grass species. Evolutionary Bioinformatics, 2016, 12: 9-21. |
| [21] | 张颖, 李峰, 刘崇怀, 樊秀彩, 孙海生, 姜建福, 张国海. 中国野生刺葡萄抗白腐病NBS-LRR类抗病基因同源序列的分离与鉴定. 中国农业科学, 2013, 46(4): 780-789. |
| ZHANG Y, LI F, LIU C H, FAN X C, SUN H S, JIANG J F, ZHANG G H. Isolation and identification of NBS-LRR resistance gene analogs sequences from Vitis davidii. Scientia Agricultura Sinica, 2013, 46(4): 780-789. (in Chinese) | |
| [22] |
MUN J H, YU H J, PARK S, PARK B S. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Molecular Genetics and Genomics, 2009, 282(6): 617-631.
doi: 10.1007/s00438-009-0492-0 |
| [23] |
LIU Y, LI D L, YANG N, ZHU X L, HAN K X, GU R, BAI J Y, WANG A X, ZHANG Y W. Genome-wide identification and analysis of CC-NBS-LRR family in response to downy mildew and black rot in Chinese cabbage. International Journal of Molecular Sciences, 2021, 22(8): 4266.
doi: 10.3390/ijms22084266 |
| [24] | 李任建, 申哲源, 李旭凯, 韩渊怀, 张宝俊. 谷子NBS-LRR类基因家族全基因组鉴定及表达分析. 河南农业科学, 2020, 49(2): 34-43. |
| LI R J, SHEN Z Y, LI X K, HAN Y H, ZHANG B J. Genome-wide identification and expression analysis of NBS-LRR gene family in Setaria italica. Journal of Henan Agricultural Sciences, 2020, 49(2): 34-43. (in Chinese) | |
| [25] |
LOUTRE C, WICKER T, TRAVELLA S, GALLI P, SCOFIELD S, FAHIMA T, FEUILLET C, KELLER B. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. The Plant Journal, 2009, 60(6): 1043-1054.
doi: 10.1111/j.1365-313X.2009.04024.x |
| [26] |
HAYASHI N, INOUE H, KATO T, FUNAO T, SHIROTA M, SHIMIZU T, KANAMORI H, YAMANE H, HAYANO-SAITO Y, MATSUMOTO T, YANO M, TAKATSUJI H. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. The Plant Journal, 2010, 64(3): 498-510.
doi: 10.1111/j.1365-313X.2010.04348.x |
| [27] |
XING L P, HU P, LIU J Q, WITEK K, ZHOU S, XU J F, ZHOU W H, GAO L, HUANG Z P, ZHANG R Q, et al. Pm21 from Haynaldia villosa encodes a CC-NBS-LRR that confers powdery mildew resistance in wheat. Molecular Plant, 2018, 11(6): 874-878.
doi: 10.1016/j.molp.2018.02.013 |
| [28] |
WANG J H, TIAN W, TAO F, WANG J J, SHANG H S, CHEN X M, XU X M, HU X P. TaRPM1 positively regulates wheat high- temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. Frontiers in Plant Science, 2020, 10: 1679.
doi: 10.3389/fpls.2019.01679 |
| [29] |
WAN H J, YUAN W, BO K L, SHEN J, PANG X, CHEN J F. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genomics, 2013, 14: 109.
doi: 10.1186/1471-2164-14-109 |
| [30] | 郝俊杰, 李磊, 王波, 秦玉红, 崔健, 王瑛, 王佩圣, 江志训, 孙吉禄, 王珍青, 岳欢, 张守才. 黄瓜白粉病抗性基因定位及候选基因分析. 中国农业科学, 2018, 51(17): 3427-3434. |
| HAO J J, LI L, WANG B, QIN Y H, CUI J, WANG Y, WANG P S, JIANG Z X, SUN J L, WANG Z Q, YUE H, ZHANG S C. Fine mapping and analysis candidate gene to powdery mildew in cucumber (Cucumis sativus L.). Scientia Agricultura Sinica, 2018, 51(17): 3427-3434. (in Chinese) | |
| [31] | ALTSCHUL S F, MADDEN T L, SCHAFFER A A, ZHANG J, ZHANG Z, MILLER W, LIPMAN D J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 1997, 25(17): 3389-3402. |
| [32] |
BIASINI M, BIENERT S, WATERHOUSE A, ARNOLD K, STUDER G, SCHMIDT T, KIEFER F, CASSARINO T G, BERTONI M, BORDOLI L, SCHWEDE T. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 2014, 42(Web Server issue): W252-W258.
doi: 10.1093/nar/gku340 |
| [33] |
HEATH M C. Hypersensitive response-related death. Plant Molecular Biology, 2000, 44(3): 321-334.
pmid: 11199391 |
| [34] |
JOSÉ-ESTANYOL M, GOMIS-RÜTH F X, PUIGDOMÈNECH P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiology and Biochemistry, 2004, 42(5): 355-365.
doi: 10.1016/j.plaphy.2004.03.009 |
| [35] |
LIU F, ZHANG X B, LU C M, ZENG X H, LI Y J, FU D H, WU G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. Journal of Experimental Botany, 2015, 66(19): 5663-5681.
doi: 10.1093/jxb/erv313 pmid: 26139823 |
| [36] |
RUSHTON P J, REINSTÄDLER A, LIPKA V, LIPPOK B, SOMSSICH I E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. The Plant Cell, 2002, 14(4): 749-762.
doi: 10.1105/tpc.010412 |
| [37] |
XIE Z, ZHANG Z L, ZOU X L, HUANG J, RUAS P, THOMPSON D, SHEN Q J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiology, 2005, 137(1): 176-189.
pmid: 15618416 |
| [38] |
BUCHEL A S, BREDERODE F T, BOL J F, LINTHORST H J M. Mutation of GT-1 binding sites in the Pr-1A promoter influences the level of inducible gene expression in vivo. Plant Molecular Biology, 1999, 40(3): 387-396.
doi: 10.1023/A:1006144505121 |
| [39] |
QU D H, SHOW P L, MIAO X L. Transcription factor ChbZIP1 from alkaliphilic microalgae Chlorella sp. BLD enhancing alkaline tolerance in transgenic Arabidopsis thaliana. International Journal of Molecular Sciences, 2021, 22(5): 2387.
doi: 10.3390/ijms22052387 |
| [40] |
FELDBRÜGGE M, SPRENGER M, HAHLBROCK K, WEISSHAAR B. PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. The Plant Journal, 1997, 11(5): 1079-1093.
doi: 10.1046/j.1365-313X.1997.11051079.x |
| [41] |
BOSTOCK R M. Signal crosstalk and induced resistance: Straddling the line between cost and benefit. Annual Review of Phytopathology, 2005, 43: 545-580.
pmid: 16078895 |
| [42] |
MUR L A J, KENTON P, ATZORN R, MIERSCH O, WASTERNACK C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology, 2006, 140(1): 249-262.
pmid: 16377744 |
| [43] |
WANG Y H, VANDENLANGENBERG K, WEN C L, WEHNER T C, WENG Y Q. QTL mapping of downy and powdery mildew resistances in PI 197088 cucumber with genotyping-by-sequencing in RIL population. Theoretical and Applied Genetics, 2018, 131(3): 597-611.
doi: 10.1007/s00122-017-3022-1 pmid: 29159421 |
| [44] |
WANG Y H, VANDENLANGENBERG K, WEHNER T C, KRAAN P A G, SUELMANN J, ZHENG X Y, OWENS K, WENG Y Q. QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theoretical and Applied Genetics, 2016, 129(8): 1493-1505.
doi: 10.1007/s00122-016-2719-x pmid: 27147071 |
| [1] | 张克坤,陈可钦,李婉平,乔浩蓉,张俊霞,刘凤之,房玉林,王海波. 灌水量对限根栽培‘阳光玫瑰’葡萄果实发育与香气物质积累的影响[J]. 中国农业科学, 2023, 56(1): 129-143. |
| [2] | 古丽旦,刘洋,李方向,成卫宁. 小麦吸浆虫小热激蛋白基因Hsp21.9的克隆及在滞育过程与温度胁迫下的表达特性[J]. 中国农业科学, 2023, 56(1): 79-89. |
| [3] | 李青林,张文涛,徐慧,孙京京. 低磷胁迫下黄瓜木质部与韧皮部汁液的代谢物变化[J]. 中国农业科学, 2022, 55(8): 1617-1629. |
| [4] | 李桂香,李秀环,郝新昌,李智文,刘峰,刘西莉. 山东省多主棒孢对三种常用杀菌剂的敏感性监测及对氟吡菌酰胺的抗性[J]. 中国农业科学, 2022, 55(7): 1359-1370. |
| [5] | 蔡苇荻,张羽,刘海燕,郑恒彪,程涛,田永超,朱艳,曹卫星,姚霞. 基于成像高光谱的小麦冠层白粉病早期监测方法[J]. 中国农业科学, 2022, 55(6): 1110-1126. |
| [6] | 郭泽西,孙大运,曲俊杰,潘凤英,刘露露,尹玲. 查尔酮合成酶基因在葡萄抗灰霉病和霜霉病中的作用[J]. 中国农业科学, 2022, 55(6): 1139-1148. |
| [7] | 冯子恒,宋莉,张少华,井宇航,段剑钊,贺利,尹飞,冯伟. 基于无人机多光谱和热红外影像信息融合的小麦白粉病监测[J]. 中国农业科学, 2022, 55(5): 890-906. |
| [8] | 束婧婷,单艳菊,姬改革,章明,屠云洁,刘一帆,巨晓军,盛中伟,唐燕飞,李华,邹剑敏. 广西麻鸡m6A甲基转移酶基因表达与肌纤维类型及成肌分化的关系[J]. 中国农业科学, 2022, 55(3): 589-601. |
| [9] | 张洁,姜长岳,王跃进. 中国野生毛葡萄转录因子VqWRKY6与VqbZIP1互作调控抗白粉病功能分析[J]. 中国农业科学, 2022, 55(23): 4626-4639. |
| [10] | 郭绍雷,许建兰,王晓俊,宿子文,张斌斌,马瑞娟,俞明亮. 桃XTH家族基因鉴定及其在桃果实贮藏过程中的表达特性[J]. 中国农业科学, 2022, 55(23): 4702-4716. |
| [11] | 郝艳,李晓颍,叶茂,刘亚婷,王天宇,王海静,张立彬,肖啸,武军凯. ‘21世纪’桃与‘久脆’桃及其杂交后代果实挥发性成分特征分析[J]. 中国农业科学, 2022, 55(22): 4487-4499. |
| [12] | 温玉霞,张坚,王琴,王靖,裴悦宏,田绍锐,樊光进,马小舟,孙现超. 本氏烟NbMBF1c的克隆、表达及在TMV侵染过程中的功能[J]. 中国农业科学, 2022, 55(18): 3543-3555. |
| [13] | 金梦娇,刘博,王抗抗,张广忠,钱万强,万方浩. 薇甘菊光能利用及叶绿素合成在不同光照强度下的响应[J]. 中国农业科学, 2022, 55(12): 2347-2359. |
| [14] | 袁景丽,郑红丽,梁先利,梅俊,余东亮,孙玉强,柯丽萍. 花青素代谢对陆地棉叶片和纤维色泽呈现的影响[J]. 中国农业科学, 2021, 54(9): 1846-1855. |
| [15] | 束婧婷,姬改革,单艳菊,章明,巨晓军,刘一帆,屠云洁,盛中伟,唐燕飞,蒋华莲,邹剑敏. IGF1-PI3K-Akt信号通路相关基因在黄羽肉鸡肌肉和肝脏中的表达[J]. 中国农业科学, 2021, 54(9): 2027-2038. |
|
||