













中国农业科学 ›› 2025, Vol. 58 ›› Issue (18): 3648-3663.doi: 10.3864/j.issn.0578-1752.2025.18.006
张淑红1( ), 高凤菊1, 武秋颖1, 纪景欣1, 张运峰1, 许可1, 谷守芹2, 范永山1(
), 高凤菊1, 武秋颖1, 纪景欣1, 张运峰1, 许可1, 谷守芹2, 范永山1( )
)
收稿日期:2025-05-18
接受日期:2025-06-15
出版日期:2025-09-18
发布日期:2025-09-18
通信作者:
联系方式:
张淑红,E-mail:zsh3535@163.com。
基金资助:
ZHANG ShuHong1( ), GAO FengJu1, WU QiuYing1, JI JingXin1, ZHANG YunFeng1, XU Ke1, GU ShouQin2, FAN YongShan1(
), GAO FengJu1, WU QiuYing1, JI JingXin1, ZHANG YunFeng1, XU Ke1, GU ShouQin2, FAN YongShan1( )
)
Received:2025-05-18
Accepted:2025-06-15
Published:2025-09-18
Online:2025-09-18
摘要:
【目的】克隆玉米大斑病菌(Setosphaeria turcica)非ACD结构域类小分子热激蛋白HSP 9/12基因,分析其在病菌发育、侵染和HT-毒素诱导过程中的表达模式。【方法】在玉米大斑病菌全基因组范围内筛选并克隆热激蛋白HSP 9/12基因,利用生物信息学方法进行编码蛋白的理化性质分析、亚细胞定位、结构预测和系统发育分析,利用RNA-seq和RT-qPCR分析HSP 9/12基因在玉米大斑病菌发育、侵染和HT-毒素诱导过程中的表达情况。【结果】从玉米大斑病菌基因组筛选并克隆到两个HSP 9/12基因,其编码蛋白分别含有99和100个氨基酸,根据分子量分别命名为StHsp10.1和StHsp10.7。理化性质分析表明,两个HSP 9/12均为亲水蛋白,亚细胞定位预测均为细胞质并含有核定位信号,无跨膜结构域和信号肽,均含有HSP9_HSP12(PF04119)结构域;StHSP10.1为酸性不稳定蛋白,StHSP10.7为碱性稳定蛋白,均以α-螺旋为主导的二级和三级结构形式存在;StHSP10.1与酿酒酵母(Saccharomyces cerevisiae)HSP12亲缘关系较近,StHSP10.7与粟酒裂殖酵母(Schizosaccharomyces pombe)HSP9亲缘关系较近。StHSP10.1在分生孢子发育期表达量最高,其次为菌丝、附着胞和侵入钉,芽管表达量较低;玉米大斑病菌接种后StHSP10.1表达量迅速上升,接种72 h时的FPKM达到接种24 h的6.37倍;HT-毒素诱导过程中的RT-qPCR分析结果表明,随着诱导时间增加,StHSP10.1在野生型菌株(WT)中相对基因表达量显著上升,诱导14、21和28 d分别为7 d的2.9、14.1和39.8倍,但在STK1基因敲除突变体(ΔSTK1)中表达量均极低;StHSP10.7在玉米大斑病菌发育阶段、侵染过程和HT-毒素诱导过程中均表达量极低。AlphaFold 3预测显示,StHSP10.1转录起始位点上游-38—-24 bp区域同时存在TATA-box、细胞分化蛋白RCD1结合位点和bZIP转录因子StbZIP11结合位点;利用STRING在线网站构建StHSP10.1蛋白质互作网络,发现2条StHSP10.1调控路径:Ras1→STK1→StbZIP11→StHSP10.1和Ras1→UBE2→CUE1→RCD1-like→StHSP10.1,推测分别在HT-毒素合成和胁迫诱导方面发挥重要作用。【结论】玉米大斑病菌HSP 9/12基因的表达模式存在显著差异,StHSP10.1是病菌发育、侵染和HT-毒素诱导过程中的关键调控基因,而StHSP10.7无调控作用。
张淑红, 高凤菊, 武秋颖, 纪景欣, 张运峰, 许可, 谷守芹, 范永山. 玉米大斑病菌热激蛋白HSP 9/12基因的克隆与表达分析[J]. 中国农业科学, 2025, 58(18): 3648-3663.
ZHANG ShuHong, GAO FengJu, WU QiuYing, JI JingXin, ZHANG YunFeng, XU Ke, GU ShouQin, FAN YongShan. Cloning and Expression Analysis of Heat Shock Protein HSP 9/12 Genes in Setosphaeria turcica[J]. Scientia Agricultura Sinica, 2025, 58(18): 3648-3663.
表1
引物序列"
| 引物名称 Primer name | 基因特异性引物序列 Gene specific primer sequence (5′-3′) | 引物名称 Primer name | 表达分析用引物序列 Sequence for gene expression analysis (5′-3′) | |
|---|---|---|---|---|
| StHSP10.1 F | ATGTCTGACTCCATGCGCAA | StHSP10.1 qF | CAGGCCTCGGAGAAGATCAC | |
| StHSP10.1 R | TTACTTCTTCTGGCCGCCAG | StHSP10.1 qR | CACGCCGGAGCTCTTGTTAG | |
| StHSP10.7 F | ATGACCTCTGCCTTCCGC | StHSP10.7 qF | CTCGGTGAAGCAATGCAACC | |
| StHSP10.7 R | TTACAACCCATTCCCATGCG | StHSP10.7 qR | CACATTGCGGTCGTAGGAGC | |
| β-tubulin qF | CAACGAAGCCTCCAACAACA | |||
| β-tubulin qR | CTCGGTGTAGTGACCCTTTGC |
表2
玉米大斑病菌HSP 9/12的理化性质和亚细胞定位预测"
| 项目Item | StHSP10.1 | StHSP10.7 |
|---|---|---|
| 氨基酸数目Number of amino acids | 99 | 100 |
| 等电点pI | 4.98 | 9.65 |
| 疏水性系数Grand average of hydropathicity | -1.06 | -1.04 |
| 不稳定系数Instability index | 44.95 | 23.63 |
| 亚细胞定位Subcellular localization | 细胞质Cytoplasm(可能性Probability 0.7207>阈值Threshold 0.4761) | 细胞质Cytoplasm(可能性Probability 0.7646>阈值Threshold 0.4761) |
| 细胞核Nucleus(可能性Probability 0.4289<阈值Threshold 0.5014) | 细胞核Nucleus(可能性Probability 0.4142<阈值Threshold 0.5014) | |
| 膜关联Membrane association:可溶Soluble(可能性Probability 0.604>阈值Threshold 0.500) | 膜关联Membrane association:外周Peripheral(可能性Probability 0.730>阈值Threshold 0.600),可溶Soluble(可能性Probability 0.660>阈值Threshold 0.500) | |
| 核定位信号Nuclear localization signal | 核定位信号Nuclear localization signal |
表4
与StHSP10.1表达正相关系数>0.9的转录因子"
| 蛋白质ID Protein ID | 相关系数 r_pearson | 相关系数P值 <BOLD>P </BOLD>val_pearson | 注释 Annotation |
|---|---|---|---|
| 178071 | 0.9620 | 6.79E-14 | PF00583乙酰转移酶(GNAT)家族Acetyltransferase (GNAT) family |
| 162963 | 0.9513 | 9.83E-13 | PF04082真菌特异性转录因子结构域Fungal specific transcription factor domain |
| 168401 | 0.9352 | 2.13E-11 | PF00397 WW结构域WW domain |
| 111166 | 0.9237 | 1.22E-10 | PF07716碱性亮氨酸拉链Basic region leucine zipper |
| 184968 | 0.9208 | 1.81E-10 | KOG2422未表征的保守蛋白Uncharacterized conserved protein |
| 39934 | 0.9204 | 1.92E-10 | KOG2177预测的E3泛素连接酶Predicted E3 ubiquitin ligase |
| 181354 | 0.9150 | 3.83E-10 | KOG3036参与细胞分化/有性发育的蛋白质Protein involved in cell differentiation/ sexual development PF04078细胞分化家族,RCD1样蛋白Cell differentiation family, RCD1-like |
| 1022253 | 0.9027 | 1.60E-09 | PF00856 SET结构域SET domain |
| 162257 | 0.9009 | 1.95E-09 | PF13523乙酰转移酶(GNAT)结构域Acetyltransferase (GNAT) domain |
表5
StHSP10.1上游顺式作用元件预测"
| 作用顺式元件cis-regulatory element | 数目Number |
|---|---|
| Short_function | 31 |
| 参与茉莉酸甲酯响应的顺式作用调控元件cis-acting regulatory element involved in the MeJA-responsiveness | 20 |
| 参与光响应性的顺式作用调控元件cis-acting regulatory element involved in light responsiveness | 9 |
| 启动子和增强子区域常见的顺式作用元件Common cis-acting element in promoter and enhancer regions | 8 |
| 参与脱落酸响应的顺式作用元件cis-acting element involved in the abscisic acid responsiveness | 7 |
| 转录起始位点上游约-30处的核心启动子元件Core promoter element around -30 of transcription start site | 7 |
| 光响应元件的一部分Part of a light responsive element | 4 |
| 光响应元件Light responsive element | 3 |
| 生长素响应元件Auxin-responsive element | 1 |
| 参与低温响应的顺式作用元件cis-acting element involved in low-temperature responsiveness | 1 |
| 参与干旱诱导性的MYB结合位点MYB binding site involved in drought-inducibility | 1 |
| MYBHv1结合位点MYBHv1 binding site | 1 |
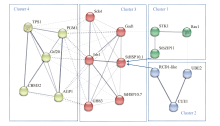
图8
StHSP10.1的蛋白质互作网络 Cluster 1:应激调节MAPK级联途径Stress-regulated MAPK cascades;Cluster 2:细胞分化基因的转录调控Transcriptional regulation of cell differentiation genes;Cluster 3:应激耐受下的HSP 9/12 Heat shock protein 9/12 under stress tolerance;Cluster 4:糖原和海藻糖代谢Glycogen and trehalose metabolism。实线表示强相互作用,虚线表示弱相互作用,线越粗相互作用越强Solid lines represent strong interactions, dashed lines represent weak interactions, and thicker lines indicate stronger interactions;箭头表示转录调控The arrows indicate transcriptional regulation;节点数Number of nodes:17;边数Number of edges:48;平均节点度Average node degree:5.65;平均局部聚类系数Average local clustering coefficient:0.613;预期边数Expected number of edges:4;PPI富集P值PPI enrichment P value:<1.0e-16"
表6
StHSP10.1蛋白质互作网络的基因信息"
| 基因名称 Gene name | 蛋白质ID Protein ID | 相关系数 r_pearson | 相关系数P值 <BOLD>P</BOLD>val_pearson | 注释 Annotation | Cluster |
|---|---|---|---|---|---|
| STK1 | 47519 | 0.4551 | 0.0254 | cd07856 STKc_Sty1_Hog1 | 1 |
| Ras1 | 169673 | 0.5297 | 0.0077 | PF00071 Ras1 | 1 |
| StbZIP11 | 155297 | 0.4830 | 0.0167 | IPR004827 bZIP | 1 |
| UBE2 | 92314 | 0.3633 | 0.0809 | PF00179泛素结合酶Ubiquitin-conjugating enzyme | 2 |
| CUE1 | 172535 | 0.7563 | 1.90E-05 | IPR040192 CUE结构域蛋白1 CUE domain-containing protein 1 | 2 |
| RCD1-like | 181354 | 0.9150 | 3.83E-10 | PF04078细胞分化家族,Rcd1样蛋白Cell differentiation family, Rcd1-like | 2 |
| Ish1 | 77830 | 0.6733 | 0.0003 | IPR018803 Ish1/Msc1-like | 3 |
| Sdo1 | 162233 | 0.9381 | 1.30E-11 | PF09377 SBDS蛋白质结构域II SBDS protein domain II | 3 |
| Gad1 | 125179 | 0.7178 | 7.85E-05 | IPR010107谷氨酸脱羧酶Glutamate decarboxylase | 3 |
| StHSP10.7 | 26210 | 0.4599 | 0.0237 | PF04119热激蛋白9/12 Heat shock protein 9/12 HSP 9_HSP12 | 3 |
| GH63 | 166579 | 0.0178 | 0.9342 | IPR004888糖苷水解酶家族63 Glycoside hydrolase family 63 | 3 |
| AGP1 | 169279 | -0.6122 | 0.0014 | IPR011834 α-葡聚糖磷酸化酶Alpha-glucan phosphorylase | 4 |
| PGM1 | 95885 | 0.2355 | 0.2678 | PTHR45955磷酸己糖异构酶Phosphohexose mutase | 4 |
| Gtf20 | 174551 | -0.4498 | 0.0273 | PF00982糖基转移酶家族20 Glycosyltransferase family 20 | 4 |
| CBM32 | 165151 | 0.2612 | 0.2175 | PF18344碳水化合物结合模块家族32 Carbohydrate binding module family 32 | 4 |
| TPS1 | 164742 | -0.4354 | 0.0334 | cd03788海藻糖-6-磷酸合成酶Trehalose-6-phosphate synthase | 4 |
| [1] |
doi: 10.1016/j.molcel.2010.08.001 pmid: 20797624 |
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
doi: 10.1007/s00114-024-01903-x pmid: 38483597 |
| [8] |
范永山, 曹志艳, 谷守芹, 董金皋. 不同诱导因素对玉米大斑病菌附着胞产生的影响. 中国农业科学, 2004, 37(5): 769-772.
|
|
|
|
| [9] |
范永山, 谷守芹, 董金皋, 董娜. MAPK途径对玉米大斑病菌 HT-毒素产生和生物学活性的调控作用. 中国农业科学, 2008, 41(1): 86-92.
|
|
|
|
| [10] |
doi: 10.1016/j.cellsig.2011.05.009 pmid: 21616144 |
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
张淑红, 张运峰, 高凤菊, 武秋颖, 许可, 李亚子, 李艳梅, 谷守芹, 范永山, 巩校东. 玉米大斑病菌小分子热激蛋白基因克隆与表达分析. 中国农业科学, 2024, 57(17): 3384-3397. doi: 10.3864/j.issn.0578-1752.2024.17.006.
|
|
|
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
张晓雅, 李学然, 薛江芝, 王小敏, 刘玉卫, 谷守芹, 董金皋. 玉米大斑病菌StSLT2基因结构与表达模式分析. 河北农业大学学报, 2019, 42(3): 32-37.
|
|
|
|
| [22] |
|
| [23] |
张运峰, 张淑红, 武秋颖, 范永山. STK1对玉米大斑病菌附着胞发育过程中糖原和脂肪积累的影响. 中国农业科学, 2017, 50(15): 2928-2935. doi: 10.3864/j.issn.0578-1752.2017.15.007.
|
|
|
|
| [24] |
doi: 10.1128/MCB.18.2.887 pmid: 9447985 |
| [25] |
|
| [26] |
张淑红, 张运峰, 高凤菊, 武秋颖, 李亚子, 许可, 范永山, 刘玉卫. 玉米大斑病菌bZIP基因家族鉴定及HT-毒素诱导过程中的表达. 微生物学报, 2025, 65(1): 283-302.
|
|
|
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
pmid: 12787503 |
| [31] |
|
| [32] |
doi: 10.1038/ng1994 pmid: 17353896 |
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
doi: 10.1096/fj.05-4184fje pmid: 16641199 |
| [1] | 滕梦鑫, 徐亚, 何静, 汪奇, 乔飞, 李敬阳, 李新国. 香蕉Ca2+-ATPase基因家族的鉴定及功能分析[J]. 中国农业科学, 2025, 58(7): 1418-1433. |
| [2] | 郑雅琴, 刘雪晴, 吴思文, 唐小燕, 杨丹妮, 汪永康, Ahmad Aftab, Khna Afrsyab, 汪承刚, 陈国户. 乌菜BcDET2的克隆及其抽薹开花调控功能[J]. 中国农业科学, 2025, 58(5): 991-1003. |
| [3] | 王慧玲, 张莹莹, 闫爱玲, 王晓玥, 刘振华, 任建成, 徐海英, 孙磊. 多组学联合解析红色玫瑰香型葡萄品种转色过程中单萜和花色苷积累规律[J]. 中国农业科学, 2025, 58(13): 2645-2662. |
| [4] | 阮桥君, 毛苗苗, 张媛媛, 林晓蓉, 陈忠正. 茶树‘英红9号’乙胺合成关键酶基因CsAlaDC转录因子的筛选[J]. 中国农业科学, 2025, 58(12): 2427-2438. |
| [5] | 吕树伟, 唐璇, 李晨. 水稻落粒性研究进展[J]. 中国农业科学, 2025, 58(1): 1-9. |
| [6] | 王程泽, 张燕, 付伟, 贾京哲, 董金皋, 申珅, 郝志敏. 玉米ACO基因家族生物信息学及表达模式分析[J]. 中国农业科学, 2024, 57(7): 1308-1318. |
| [7] | 张博文, 赵丽雯, 徐璐, 李盼, 曾凡力, 孟亚南, 董金皋. 玉米大斑病菌细胞周期蛋白依赖性激酶结构亚基StCks1鉴定及其基因功能分析[J]. 中国农业科学, 2024, 57(5): 900-908. |
| [8] | 张淑红, 张运峰, 高凤菊, 武秋颖, 许可, 李亚子, 李艳梅, 谷守芹, 范永山, 巩校东. 玉米大斑病菌小分子热激蛋白基因克隆与表达分析[J]. 中国农业科学, 2024, 57(17): 3384-3397. |
| [9] | 葛天成, 尹飞, 胡琼波, 彭争科, 李振宇. MBF2转录调控小菜蛾谷胱甘肽S-转移酶代谢氯虫苯甲酰胺的功能[J]. 中国农业科学, 2023, 56(4): 665-673. |
| [10] | 王壮壮, 董邵云, 周琪, 苗晗, 刘小萍, 徐奎鹏, 顾兴芳, 张圣平. 黄瓜果实维生素C合成关键基因克隆与分析[J]. 中国农业科学, 2023, 56(3): 508-518. |
| [11] | 邹金鹏, 岳浩峰, 李海笑, 刘峥, 刘宁, 曹志艳, 董金皋. 基于非靶向代谢组学分析StLAC2和StLAC6差异影响玉米大斑病菌的机制[J]. 中国农业科学, 2023, 56(16): 3110-3223. |
| [12] | 古丽旦,刘洋,李方向,成卫宁. 小麦吸浆虫小热激蛋白基因Hsp21.9的克隆及在滞育过程与温度胁迫下的表达特性[J]. 中国农业科学, 2023, 56(1): 79-89. |
| [13] | 李雨泽,朱嘉伟,林蔚,蓝茉莹,夏黎明,张艺粒,罗聪,黄桂香,何新华. 香水柠檬RHF2A的克隆与互作蛋白的筛选[J]. 中国农业科学, 2022, 55(24): 4912-4926. |
| [14] | 张洁, 姜长岳, 王跃进. 中国野生毛葡萄转录因子VqWRKY6与VqbZIP1互作调控抗白粉病功能分析[J]. 中国农业科学, 2022, 55(23): 4626-4639. |
| [15] | 郝玉彬,李海笑,张赛,刘宁,刘英姿,曹志艳,董金皋. 玉米大斑病菌小柱孢酮脱水酶StSCD家族鉴定及其功能分析[J]. 中国农业科学, 2022, 55(16): 3134-3143. |
|
||