













中国农业科学 ›› 2022, Vol. 55 ›› Issue (13): 2552-2561.doi: 10.3864/j.issn.0578-1752.2022.13.006
收稿日期:2021-12-17
接受日期:2022-01-18
出版日期:2022-07-01
发布日期:2022-07-08
联系方式:
渠成,E-mail: quch1990@sina.com。
基金资助:
QU Cheng( ),WANG Ran,LI FengQi,LUO Chen(
),WANG Ran,LI FengQi,LUO Chen( )
)
Received:2021-12-17
Accepted:2022-01-18
Published:2022-07-01
Online:2022-07-08
摘要:
【目的】烟粉虱(Bemisia tabaci)为世界性分布的重要农林入侵害虫,寄主植物种类众多,但对不同寄主植物存在嗜性差异,而味觉受体(gustatory receptor,GR)基因在其取食选择等行为中发挥重要作用。本研究通过克隆烟粉虱味觉受体GR1和GR2基因,明确其在烟粉虱不同发育期和成虫不同组织中的表达特性,为基因功能的深入研究提供依据。【方法】从烟粉虱MED隐种成虫头部转录组中筛选获得味觉受体基因序列,利用巢式PCR技术克隆获得两个味觉受体基因的完整开放阅读框(open reading frame,ORF)序列,两个基因分别命名为BtabGR1和BtabGR2,运用生物信息学软件对其编码氨基酸序列特征、结构域等信息进行预测,基于邻接法构建BtabGR1、BtabGR2与其他半翅目昆虫味觉受体基因的系统进化树,并利用实时荧光定量PCR方法分析两个基因在烟粉虱不同发育期(卵、1—4龄若虫和雌、雄成虫)和雌、雄成虫不同组织(头、胸、腹、足)中的表达情况。【结果】克隆获得BtabGR1和BtabGR2基因序列(GenBank登录号:OL845904和OL845905),完整开放阅读框为1 287和1 344 bp,分别编码428和447个氨基酸,预测蛋白分子量为 48.54和51.50 kD,理论等电点为8.85和8.74,具有4个和6个跨膜结构域。氨基酸序列比对和系统进化分析结果显示,BtabGR1 和BtabGR2与其他半翅目昆虫GR28b和GR43a-like基因亲缘关系较近。发育期表达谱分析表明,BtabGR1和BtabGR2在烟粉虱的不同发育历期均有表达,其中BtabGR1在成虫中表达量高于其他发育期,而BtabGR2在卵中的表达量最高;组织表达谱结果表明,BtabGR1和BtabGR2在烟粉虱雌、雄成虫不同组织中均有表达,且均在成虫头部高表达。【结论】BtabGR1和BtabGR2具有昆虫味觉受体基因的典型特征,二者均在烟粉虱成虫头部高表达,推测两基因在烟粉虱寄主植物适应过程中发挥重要作用,可作为烟粉虱新型防控措施的潜在靶标。
渠成,王然,李峰奇,罗晨. 烟粉虱味觉受体基因BtabGR1和BtabGR2的克隆与表达模式分析[J]. 中国农业科学, 2022, 55(13): 2552-2561.
QU Cheng,WANG Ran,LI FengQi,LUO Chen. Cloning and Expression Profiling of Gustatory Receptor Genes BtabGR1 and BtabGR2 in Bemisia tabaci[J]. Scientia Agricultura Sinica, 2022, 55(13): 2552-2561.
表1
引物序列"
| 引物 Primer | 引物序列 Primer sequence (5′-3′) | 引物用途 Usage of primers |
|---|---|---|
| GR1-1441-F | CCTCTAGTTGTTCTTCTATCG | 开放阅读框扩增 Amplification of ORF |
| GR1-1441-R | TGTCTGAATCATTTTTTATCC | |
| GR1-ORF-F | ATGCAATTCCACTTGATTCCGACAT | |
| GR1-ORF-R | TTATGAAGTTGAATTATGTGAAAAT | |
| GR2-1495-F | AAAAAGGAAACATGAAAAAAA | |
| GR2-1495-R | GAACTGTACGTATAAGAGCTG | |
| GR2-ORF-F | ATGAAATTTAAAAACCGCACATTCT | |
| GR2-ORF-R | TTAAGACACCATGTTTTGTTTTTCC | |
| GR1-117-F | ACCTAAACAGTCCCAAGTGCT | 实时荧光定量PCR qRT-PCR |
| GR1-117-R | CTTCGAACCCGTCACACGAA | |
| GR2-181-F | AAAAGACAAGCTGGTCCCCG | |
| GR1-181-R | GTCGGCTAAACTGTGGGTGT | |
| EF1α-110-F | TAGCCTTGTGCCAATTTCCG | |
| EF1α-110-R | CCTTCAGCATTACCGTCC | |
| RPL29-144-F | TCGGAAAATTACCGTGAG | |
| RPL29-144-R | GAACTTGTGATCTACTCCTCTCGTG |

图4
BtabGR1、BtabGR2与其他半翅目昆虫味觉受体氨基酸序列比对 SflaGR28b:甘蔗黄蚜Sipha flava (XP_025417386.1);AgosGR28b:棉蚜Aphis gossypii (XP_027840408.1);ApisGR28b、ApisGR43a-like:豌豆蚜Acyrthosiphon pisum (XP_008178128.1, XP_003244120.4);MperGR43a-like:桃蚜Myzus persicae (XP_022165389.1);MsacGR43a-like:高粱蚜Melanaphis sacchari (XP_025195577.1);Consensus:共有序列Consensus sequence。黑色区域代表相似度100%,粉红色区域代表相似度大于75%,浅蓝色区域代表相似度大于50% Black areas indicate 100% similarity, pink areas indicate more than 75% similarity, blue areas indicate more than 50% similarity"

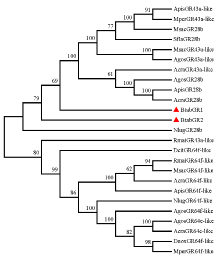
图5
基于氨基酸序列构建的BtabGR1、BtabGR2与其他半翅目昆虫味觉受体的系统进化树 GR蛋白来源物种及GenBank登录号Origin species of GRs and their GenBank accession numbers。BtabGR1、BtabGR2:烟粉虱Bemisia tabaci (OL845904, OL845905);ApisGR43a-like、ApisGR28b、ApisGR64f-like:豌豆蚜Acyrthosiphon pisum (XP_003244120.4, XP_008178128.1, XP_016660313.2); AcraGR43a-like、AcraGR64e-like、AcraGR64f-like、AcraGR28b:豆蚜Aphis craccivora (KAF0773920.1, KAF0765871.1, KAF0765661.1, KAF0749339.1);AgosGR28b、AgosGR43a-like、AgosGR64f-like、AgosGR64e-like:棉蚜Aphis gossypii (XP_027840408.1, XP_027840395.1, XP_027847490.1, XP_027847461.1);DcitGR64f-like:柑橘木虱Diaphorina citri (XP_017300309.2);DnoxGR64f-like:麦双尾蚜Diuraphis noxia (XP_015369418.1); MperGR43a-like、MperGR64f-like:桃蚜Myzus persicae (XP_022165389.1, XP_016660313.2);MsacGR43a-like、MsacGR64f-like、MsacGR28b:高粱蚜Melanaphis sacchari (XP_025195577.1, XP_025204513.1, XP_025202779.1);NlugGR28b、NlugGR64f-like:褐飞虱Nilaparvata lugens (XP_039287957.1, XP_022186608.2);RmaiGR43a-like、RmaiGR64f-like:玉米蚜Rhopalosiphum maidis (XP_026807230.1, XP_026822156.1);SflaGR28b:甘蔗黄蚜Sipha flava (XP_025417386.1)"

| [1] |
DE BARRO P J, DRIVER F. Use of RAPD PCR to distinguish the B biotype from other biotypes of Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae). Australian Journal of Entomology, 1997, 36(2): 149-152.
doi: 10.1111/j.1440-6055.1997.tb01447.x |
| [2] |
DE BARRO P J, LIU S S, BOYKIN L M, DINSDALE A B. Bemisia tabaci: A statement of species status. Annual Review of Entomology, 2011, 56: 1-19.
doi: 10.1146/annurev-ento-112408-085504 |
| [3] |
PAN H P, CHU D, YAN W Q, SU Q, LIU B M, WANG S L, WU Q J, XIE W, JIAO X G, LI R M, et al. Rapid spread of tomato yellow leaf curl virus in China is aided differentially by two invasive whiteflies. PLoS ONE, 2012, 7(4): e34817.
doi: 10.1371/journal.pone.0034817 |
| [4] |
TANG X, SHI X B, ZHANG D Y, LI F, YAN F, ZHANG Y J, LIU Y, ZHOU X G. Detection and epidemic dynamic of ToCV and CCYV with Bemisia tabaci and weed in Hainan of China. Virology Journal, 2017, 14: 169.
doi: 10.1186/s12985-017-0833-2 |
| [5] |
CHU D, ZHANG Y J, BROWN J K, CONG B, XU B Y, WU Q J, ZHU G R. The introduction of the exotic Q biotype of Bemisia tabaci from the Mediterranean region into China on ornamental crops. Florida Entomologist, 2006, 89(2): 168-174.
doi: 10.1653/0015-4040(2006)89[168:TIOTEQ]2.0.CO;2 |
| [6] |
HU J, DE BARRO P, ZHAO H, WANG J, NARDI F, LIU S S. An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS ONE, 2011, 6(1): e16061.
doi: 10.1371/journal.pone.0016061 |
| [7] |
TANG X T, CAI L, SHEN Y, XU L L, DU Y Z. Competitive displacement between Bemisia tabaci MEAM1 and MED and evidence for multiple invasions of MED. Insects, 2020, 11(1): 35.
doi: 10.3390/insects11010035 |
| [8] |
XUE Y T, LIN C T, WANG Y Z, ZHANG Y B, JI L Q. Ecological niche complexity of invasive and native cryptic species of the Bemisia tabaci species complex in China. Journal of Pest Science, 2022, 95: 1245-1259.
doi: 10.1007/s10340-021-01450-8 |
| [9] |
OLIVEIRA M R V, HENNEBERRY T J, ANDERSON P. History, current status, and collaborative research projects for Bemisia tabaci. Crop Protection, 2001, 20(9): 709-723.
doi: 10.1016/S0261-2194(01)00108-9 |
| [10] | 张永军, 梁革梅, 倪云霞, 吴孔明, 郭予元. 烟粉虱成虫对不同寄主植物的选择性. 植物保护, 2003, 29(2): 20-22. |
| ZHANG Y J, LIANG G M, NI Y X, WU K M, GUO Y Y. Selection of the adult of Bemisia tabaci (Gennadius) to different host plants. Plant Protection, 2003, 29(2): 20-22. (in Chinese) | |
| [11] | 周福才, 黄振, 王勇, 李传明, 祝树德. 烟粉虱(Bemisia tabaci)的寄主选择性. 生态学报, 2008, 28(8): 3825-3831. |
| ZHOU F C, HUANG Z, WANG Y, LI C M, ZHU S D. Host plant selection of Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae). Acta Ecologica Sinica, 2008, 28(8): 3825-3831. (in Chinese) | |
| [12] | WAN F H, ZHANG G F, LIU S S, LUO C, CHU D, ZHANG Y J, ZANG L S, JIU M, LÜ Z Q, CUI X H, et al. Invasive mechanism and management strategy of Bemisia tabaci (Gennadius) biotype B:Progress report of 973 Program on invasive alien species in China. Science in China Series C-Life Sciences, 2009, 52(1): 88-95. |
| [13] |
LI Y F, ZHONG S T, QIN Y C, ZHANG S Q, GAO Z L, DANG Z G, PAN W L. Identification of plant chemicals attracting and repelling whiteflies. Arthropod-Plant Interactions, 2014, 8(3): 183-190.
doi: 10.1007/s11829-014-9302-7 |
| [14] |
SACCHETTI P, ROSSI E, BELLINI L, VERNIERI P L, CIONI P L, FLAMINI G. Volatile organic compounds emitted by bottlebrush species affect the behaviour of the sweet potato whitefly. Arthropod- Plant Interactions, 2015, 9(4): 393-403.
doi: 10.1007/s11829-015-9382-z |
| [15] |
SADEH D, NITZAN N, SHACHTER A, CHAIMOVITSH D, DUDAI N, GHANIM M. Whitefly attraction to rosemary (Rosmarinus officinialis L.) is associated with volatile composition and quantity. PLoS ONE, 2017, 12(5): e0177483.
doi: 10.1371/journal.pone.0177483 |
| [16] |
OZAKI K, RYUDA M, YAMADA A, UTOGUCHI A, ISHIMOTO H, CALAS D, MARION-POLL F, TANIMURA T, YOSHIKAWA H. A gustatory receptor involved in host plant recognition for oviposition of a swallowtail butterfly. Nature Communications, 2011, 2: 542.
doi: 10.1038/ncomms1548 |
| [17] |
BRISCOE A D, MACIAS-MUNOZ A, KOZAK K M, WALTERS J R, YUAN F R, JAMIE G A, MARTIN S H, DASMAHAPSATRA K K, FERGUSON L C, MALLET J, JACQUIN-JOLY E, JIGGINS C D. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genetics, 2013, 9(7): e1003620.
doi: 10.1371/journal.pgen.1003620 |
| [18] | SCHOONHOVEN L M, VAN LOON J J A. An inventory of taste in caterpillars: Each species its own key. Acta Zoologica Academiae Scientiarum Hungaricae, 2002, 48(Suppl. 1): 215-263. |
| [19] |
LIU X L, YAN Q, YANG Y L, HOU W, MIAO C L, PENG Y C, DONG S L. A gustatory receptor GR8 tunes specifically to D-fructose in the common cutworm Spodoptera litura. Insects, 2019, 10(9): 272.
doi: 10.3390/insects10090272 |
| [20] |
CLYNE P J, WARR C G, CARLSON J R. Candidate taste receptors in Drosophila. Science, 2000, 287(5459): 1830-1834.
doi: 10.1126/science.287.5459.1830 |
| [21] |
XU W, ZHANG H J, ANDERSON A. A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. Journal of Chemical Ecology, 2012, 38(12): 1513-1520.
doi: 10.1007/s10886-012-0221-8 |
| [22] |
HE P, ENGSONTIA P, CHEN G L, YIN Q, WANG J, LU X, ZHANG Y N, LI Z Q, HE M. Molecular characterization and evolution of a chemosensory receptor gene family in three notorious rice planthoppers, Nilaparvata lugens, Sogatella furcifera and Laodelphax striatellus, based on genome and transcriptome analyses. Pest Management Science, 2018, 74(9): 2156-2167.
doi: 10.1002/ps.4912 |
| [23] |
KANG K, YANG P, CHEN L E, PANG R, YU L J, ZHOU W W, ZHU Z R, ZHANG W Q. Identification of putative fecundity-related gustatory receptor genes in the brown planthopper Nilaparvata lugens. BMC Genomics, 2018, 19(1): 970.
doi: 10.1186/s12864-018-5391-5 |
| [24] |
SMAGJA C, SHI P, BUTLIN R K, ROBERTSON H M. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Molecular Biology and Evolution, 2009, 26(9): 2073-2086.
doi: 10.1093/molbev/msp116 |
| [25] |
WANG R, LI F Q, ZHANG W, ZHANG X M, QU C, TETREAU G, SUN L J, LUO C, ZHOU J J. Identification and expression profile analysis of odorant binding protein and chemosensory protein genes in Bemisia tabaci MED by head transcriptome. PLoS ONE, 2017, 12(2): e0171739.
doi: 10.1371/journal.pone.0171739 |
| [26] |
ZENG Y, YANG Y T, WU Q J, WANG S L, XIE W, ZHANG Y J. Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Science, 2019, 26(4): 620-634.
doi: 10.1111/1744-7917.12576 |
| [27] |
LI R M, XIE W, WANG S L, WU Q J, YANG N N, YANG X, PAN H P, ZHOU X M, BAI L Y, XU B Y, ZHOU X G, ZHANG Y Y. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE, 2013, 8(1): e53006.
doi: 10.1371/journal.pone.0053006 |
| [28] | SATO K, TANAKA K, TOUHARA K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(28): 11680-11685. |
| [29] |
MANG D Z, SHU M, ENDO H, YOSHIZAWA Y, NAGATA S, KIKUTA S, SATO R. Expression of a sugar clade gustatory receptor, BmGr6, in the oral sensory organs, midgut, and central nervous system of larvae of the silkworm Bombyx mori. Insect Biochemistry and Molecular Biology, 2016, 70: 85-98.
doi: 10.1016/j.ibmb.2015.12.008 |
| [30] |
JIANG X J, NING C, GUO H, JIA Y Y, HUANG L Q, QU M J, WANG C Z. A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect Biochemistry and Molecular Biology, 2015, 60: 39-46.
doi: 10.1016/j.ibmb.2015.03.002 |
| [31] |
MIYAMOTO T, SLONE J, SONG X Y, AMREIN H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell, 2012, 151(5): 1113-1125.
doi: 10.1016/j.cell.2012.10.024 |
| [32] | SANG J, RIMAL S, LEE Y. Gustatory receptor 28b is necessary for avoiding saponin in Drosophila melanogaster. EMBO Reports, 2019, 20(2): e47328. |
| [33] |
XIANG Y, YUAN Q, VOGT N, LOOGER L L, JAN L Y, JAN Y N. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature, 2010, 468(7326): 921-926.
doi: 10.1038/nature09576 |
| [34] |
NI L N, BRONK P, CHANG E C, LOWELL A M, FLAM J O, PANZANO V C, THEOBALD D L, GRIFFITH L C, GARRITY P A. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature, 2013, 500(7464): 580-584.
doi: 10.1038/nature12390 |
| [35] | 刘一鹏, 刘杨, 杨婷, 桂富荣, 王桂荣. 小菜蛾普通气味受体基因PxylOR9的鉴定及功能研究. 昆虫学报, 2015, 58(5): 507-515. |
| LIU Y P, LIU Y, YANG T, GUI F R, WANG G R. Identification and characterization of a general odorant receptor gene PxylOR9 in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Acta Entomologica Sinica, 2015, 58(5): 507-515. (in Chinese) | |
| [36] |
SAINA M, BUSENGDAL H, SINIGAGLIA C, PETRONE L, OLIVERI P, RENTZSCH F, BENTON R. A cnidarian homologue of an insect gustatory receptor functions in developmental body patterning. Nature Communications, 2015, 6: 6243.
doi: 10.1038/ncomms7243 |
| [37] |
ABDEL-LATIEF M. A family of chemoreceptors in Tribolium castaneum (Tenebrionidae: Coleoptera). PLoS ONE, 2007, 2(12): e1319.
doi: 10.1371/journal.pone.0001319 |
| [38] |
XU W, PAPANICOLAOU A, LIU N Y, DONG S L, ANDERSON A. Chemosensory receptor genes in the oriental tobacco budworm Helicoverpa assulta. Insect Molecular Biology, 2015, 24(2): 253-263.
doi: 10.1111/imb.12153 |
| [39] |
AHMED T, ZHANG T T, WANG Z Y, HE K L, BAI S X. Gene set of chemosensory receptors in the polyembryonic endoparasitoid Macrocentrus cingulum. Scientific Reports, 2016, 6: 24078.
doi: 10.1038/srep24078 |
| [40] | YI J K, YANG S, WANG S, WANG J, ZHANG X X, LIU Y, XI J H. Identification of candidate chemosensory receptors in the antennal transcriptome of the large black chafer Holotrichia parallela Motschulsky (Coleoptera: Scarabaeidae). Comparative Biochemistry and Physiology. Part D, Genomics and Proteomics, 2018, 28: 63-71. |
| [41] |
SUN L, ZHANG Y N, QIAN J L, KANG K, ZHANG X Q, DENG J D, TANG Y P, CHEN C, HANSEN L, XU T, ZHANG Q H, ZHANG L W. Identification and expression patterns of Anoplophora chinensis (Forster) chemosensory receptor genes from the antennal transcriptome. Frontiers in Physiology, 2018, 9: 90.
doi: 10.3389/fphys.2018.00090 |
| [1] | 张淑红, 张运峰, 高凤菊, 武秋颖, 许可, 李亚子, 李艳梅, 谷守芹, 范永山, 巩校东. 玉米大斑病菌小分子热激蛋白基因克隆与表达分析[J]. 中国农业科学, 2024, 57(17): 3384-3397. |
| [2] | 王愿, 都梦丹, 李正刚, 佘小漫, 于琳, 蓝国兵, 丁善文, 何自福, 汤亚飞. 广东省番茄白尖曲叶病病原鉴定及其致病性分析[J]. 中国农业科学, 2024, 57(12): 2350-2363. |
| [3] | 王壮壮, 董邵云, 周琪, 苗晗, 刘小萍, 徐奎鹏, 顾兴芳, 张圣平. 黄瓜果实维生素C合成关键基因克隆与分析[J]. 中国农业科学, 2023, 56(3): 508-518. |
| [4] | 黄玉萱, 沈忱, 鞠佳菲, 杨磊, 罗光华, 方继朝. 二化螟味觉受体基因鉴定、克隆与表达模式分析[J]. 中国农业科学, 2023, 56(13): 2504-2517. |
| [5] | 古丽旦,刘洋,李方向,成卫宁. 小麦吸浆虫小热激蛋白基因Hsp21.9的克隆及在滞育过程与温度胁迫下的表达特性[J]. 中国农业科学, 2023, 56(1): 79-89. |
| [6] | 李雨泽,朱嘉伟,林蔚,蓝茉莹,夏黎明,张艺粒,罗聪,黄桂香,何新华. 香水柠檬RHF2A的克隆与互作蛋白的筛选[J]. 中国农业科学, 2022, 55(24): 4912-4926. |
| [7] | 张莉,张楠,江虎强,吴帆,李红亮. 中华蜜蜂NPC2基因家族克隆及表达模式分析[J]. 中国农业科学, 2022, 55(12): 2461-2471. |
| [8] | 郑海霞,高玉林,张方梅,杨超霞,蒋健,朱勋,张云慧,李祥瑞. 马铃薯甲虫热激蛋白基因Ld-hsp70的克隆及温度胁迫下的表达特性[J]. 中国农业科学, 2021, 54(6): 1163-1175. |
| [9] | 张璐,宗亚奇,徐维华,韩蕾,孙浈育,陈朝晖,陈松利,张凯,程杰山,唐美玲,张洪霞,宋志忠. 葡萄Fe-S簇装配基因的鉴定、克隆和表达特征分析[J]. 中国农业科学, 2021, 54(23): 5068-5082. |
| [10] | 谭永安,姜义平,赵静,肖留斌. 绿盲蝽G蛋白偶联受体激酶2基因(AlGRK2)的表达分析及在绿盲蝽生长发育中的功能[J]. 中国农业科学, 2021, 54(22): 4813-4825. |
| [11] | 叶方婷,潘鑫峰,毛志君,李兆伟,范凯. 睡莲转录因子bZIP家族的分子进化以及功能分析[J]. 中国农业科学, 2021, 54(21): 4694-4708. |
| [12] | 王娜,赵资博,高琼,何守朴,马晨辉,彭振,杜雄明. 陆地棉盐胁迫应答基因GhPEAMT1的克隆及功能分析[J]. 中国农业科学, 2021, 54(2): 248-260. |
| [13] | 何云川,王新谱,洪波,张婷婷,周雪飞,贾彦霞. 四种杀虫剂LC25对Q型烟粉虱成虫取食行为的影响[J]. 中国农业科学, 2021, 54(2): 324-333. |
| [14] | 赵付枚,王爽,田雨婷,乔奇,王永江,张德胜,张振臣. 甘薯病毒病发生关键因素研究[J]. 中国农业科学, 2021, 54(15): 3232-3240. |
| [15] | 谭永安,赵旭东,姜义平,赵静,肖留斌,郝德君. 绿盲蝽雷帕霉素靶蛋白的克隆、抗体制备及在蜕皮激素诱导下的应答[J]. 中国农业科学, 2021, 54(10): 2118-2131. |
|
||