













中国农业科学 ›› 2023, Vol. 56 ›› Issue (19): 3814-3828.doi: 10.3864/j.issn.0578-1752.2023.19.009
张昕( ), 杨星宇, 张超然, 张冲, 郑海霞, 张仙红(
), 杨星宇, 张超然, 张冲, 郑海霞, 张仙红( )
)
收稿日期:2023-06-19
接受日期:2023-07-03
出版日期:2023-10-01
发布日期:2023-10-08
通信作者:
联系方式:
张昕,E-mail:sxauzx2018@163.com。
基金资助:
ZHANG Xin( ), YANG XingYu, ZHANG ChaoRan, ZHANG Chong, ZHENG HaiXia, ZHANG XianHong(
), YANG XingYu, ZHANG ChaoRan, ZHANG Chong, ZHENG HaiXia, ZHANG XianHong( )
)
Received:2023-06-19
Accepted:2023-07-03
Published:2023-10-01
Online:2023-10-08
摘要:
【目的】鉴定绿豆象(Callosobruchus chinensis)热激蛋白(heat shock protein,HSP)超家族基因成员,明确高、低温胁迫后HSP基因在绿豆象中的表达变化,为深入挖掘HSP基因功能提供理论依据。【方法】从Insect Base 2.0下载不同昆虫HSP基因的CDS和蛋白序列,并以此为参考在绿豆象全长转录组测序数据库中进行本地BLASTp和tBLASTn比对搜索,同时结合HMMER和关键词两种方法再次筛选目标序列,最终完成搜索结果的汇总。利用CDD、MEGA、ProtParam等在线分析工具对绿豆象HSP超家族基因进行生物信息学分析。根据绿豆象成虫高、低温转录组测序数据筛选出7个候选CcHsp,采用实时荧光定量PCR(qRT-PCR)技术比较分析其在绿豆象不同虫态(幼虫、蛹、成虫)及不同温度胁迫下的表达特性。【结果】共鉴定出31个HSP基因,其中包括3个HSP90、8个HSP70、8个HSP60和12个sHSP(small HSP)。理化性质分析显示CcHsp编码的蛋白质包含159—776个氨基酸残基(aa),分子量介于18.4—88.9 kDa,理论等电点为4.95—9.17。亚细胞定位结果显示多数CcHsp定位于细胞质中,少数基因定位于线粒体基质、内质网和细胞核。系统发育分析表明绿豆象热激蛋白不同家族成员与其他昆虫的热激蛋白进化关系较近,显示了其在进化上的保守性。qRT-PCR分析发现,不同虫态在不同温度胁迫后7个候选CcHsp差异表达。CcHsp20.102经高温胁迫后在雌、雄成虫体内的表达量分别上调1 000和500倍;CcHsp70-5经高温胁迫后在雌、雄成虫体内的表达量分别上调500和450倍;幼虫经高、低温胁迫后,CcHsp19.855和CcHsp70-5表达差异显著。【结论】通过绿豆象全长转录组测序数据共鉴定出31个完整的热激蛋白超家族基因成员,分为4个亚家族,家族间蛋白结构、保守结构域和基因表达特征存在差异。CcHsp20.102和CcHsp70-5可能在成虫抵御高温胁迫中行使重要功能,而幼虫的高温耐受性可能与CcHsp19.855和CcHsp70-5的表达差异有关。
张昕, 杨星宇, 张超然, 张冲, 郑海霞, 张仙红. 绿豆象热激蛋白超家族基因的鉴定及表达分析[J]. 中国农业科学, 2023, 56(19): 3814-3828.
ZHANG Xin, YANG XingYu, ZHANG ChaoRan, ZHANG Chong, ZHENG HaiXia, ZHANG XianHong. Identification and Expression Analysis of Heat Shock Protein Superfamily Genes in Callosobruchus chinensis[J]. Scientia Agricultura Sinica, 2023, 56(19): 3814-3828.
表1
qRT-PCR引物信息及结果分析"
| 基因名 Gene name | 引物序列 Primer sequence (5′ to 3′) | 产物长度 Product length (bp) | 标准曲线 Standard curve (Y=) | 扩增效率Amplification efficacy (%) | 相关系数 Correlation coefficient |
|---|---|---|---|---|---|
| CcHsp90-2 | F: TAATCAGTCGTTGAAGCAG R: TACATTGCTGTTGCGATTC | 103 | -3.276*log2X+15.55 | 102.0 | 0.992 |
| CcHsp70-5 | F: ACAGATTCCGTGTTTTGA R: GCTAAAACTCAGTCTATTCG | 109 | -3.379*log2X+14.43 | 97.7 | 0.987 |
| CcHsp70-6 | F: CGACAGTGTAAACAACTCTAGC R: GAATGTCACAGCAGATACTACTA | 160 | -3.184*log2X+22.40 | 106.1 | 0.998 |
| CcHsp60-3 | F: GCATAATGGGTCTGCGTCG R: AGCATAGCCGAGGTTGGA | 116 | -3.178*log2X+19.36 | 106.4 | 0.998 |
| CcHsp20.102 | F: TGCTAGTGTCAGTGTTCAAC R: ACATACAAAATCAACCTTCC | 126 | -3.146*log2X+24.81 | 107.9 | 0.994 |
| CcHsp19.855 | F: GAGCGACTTTTCGTATCAA R: TCCAACACCAATCTTTCAT | 175 | -3.197*log2X+20.01 | 105.5 | 0.999 |
| CcHsp18.572 | F: CGTTTTTTGACTGGAATAC R: TGGATAGAAATACCTGCTT | 144 | -3.362*log2X+20.68 | 98.4 | 0.999 |
| β-tubulin | F: CTTCAGAGGCAGGATGTC R: TCCCCTTGGTGGAATGTC | 133 | -3.269*log2X+16.20 | 102.3 | 0.997 |
表2
绿豆象HSP超家族基因编码蛋白质的理化性质分析"
| 基因名 Gene name | 编码数 CDS number | 氨基酸数 AA number | 分子量 MW (kDa) | 等电点 PI | 不稳定指数 II | 脂肪指数 AI | 亲水性平均值 GH | 亚细胞定位 SL |
|---|---|---|---|---|---|---|---|---|
| CcHsp18.444 | 492 | 163 | 18.444 | 7.75 | 52.21 | 71.04 | -0.942 | 细胞质Cytoplasm |
| CcHsp18.572 | 480 | 159 | 18.572 | 6.45 | 39.20 | 83.96 | -0.686 | 细胞质、细胞核 Cytoplasm, nucleus |
| CcHsp19.121 | 504 | 167 | 19.121 | 8.79 | 53.82 | 73.41 | -0.863 | 细胞质Cytoplasm |
| CcHsp19.855 | 525 | 174 | 19.855 | 6.96 | 43.56 | 78.39 | -0.690 | 细胞质Cytoplasm |
| CcHsp20.102 | 531 | 176 | 20.102 | 6.97 | 48.14 | 76.42 | -0.657 | 细胞质Cytoplasm |
| CcHsp20.103 | 534 | 177 | 20.103 | 6.30 | 59.44 | 84.24 | -0.541 | 细胞质Cytoplasm |
| CcHsp20.956 | 533 | 176 | 20.956 | 6.24 | 62.18 | 77.68 | -0.658 | 细胞质Cytoplasm |
| CcHsp21.032 | 573 | 190 | 21.032 | 7.59 | 56.39 | 67.58 | -0.552 | 线粒体基质 Mitochondrial matrix |
| CcHsp21.936 | 585 | 194 | 21.936 | 8.34 | 57.23 | 85.82 | -0.730 | 细胞质、细胞核 Cytoplasm, nucleus |
| CcHsp22.154 | 591 | 196 | 22.154 | 6.18 | 56.98 | 89.90 | -0.610 | 细胞质Cytoplasm |
| CcHsp22.763 | 606 | 201 | 22.763 | 7.90 | 65.18 | 85.72 | -0.544 | 细胞质Cytoplasm |
| CcHsp24.313 | 600 | 199 | 24.313 | 7.94 | 51.07 | 62.61 | -0.957 | 细胞质Cytoplasm |
| CcHsp60-1 | 1605 | 534 | 57.326 | 6.36 | 33.89 | 111.59 | 0.078 | 细胞质Cytoplasm |
| CcHsp60-2 | 1605 | 534 | 57.594 | 5.74 | 35.65 | 101.78 | -0.063 | 细胞核Nucleus |
| CcHsp60-3 | 1593 | 530 | 57.978 | 6.84 | 28.57 | 101.94 | -0.122 | 细胞质Cytoplasm |
| CcHsp60-4 | 1626 | 541 | 58.777 | 5.84 | 40.52 | 104.21 | -0.032 | 细胞质Cytoplasm |
| CcHsp60-5 | 1623 | 540 | 59.011 | 7.49 | 32.25 | 100.07 | -0.089 | 细胞质Cytoplasm |
| CcHsp60-6 | 1593 | 530 | 60.038 | 6.74 | 32.45 | 104.41 | -0.002 | 线粒体基质 Mitochondrial matrix |
| CcHsp60-7 | 1656 | 551 | 60.969 | 5.98 | 44.86 | 96.46 | -0.230 | 细胞质Cytoplasm |
| CcHsp60-8 | 1725 | 574 | 61.300 | 5.44 | 33.69 | 95.96 | -0.170 | 线粒体基质 Mitochondrial matrix |
| CcHsp70-1 | 1755 | 584 | 64.129 | 9.17 | 37.20 | 86.68 | -0.346 | 细胞质Cytoplasm |
| CcHsp70-2 | 1767 | 588 | 64.938 | 5.66 | 21.45 | 89.20 | -0.383 | 内质网 Endoplasmic reticulum |
| CcHsp70-3 | 1887 | 628 | 69.211 | 5.55 | 33.47 | 83.73 | -0.429 | 细胞质Cytoplasm |
| CcHsp70-4 | 1905 | 634 | 69.864 | 4.97 | 25.51 | 84.89 | -0.496 | 内质网 Endoplasmic reticulum |
| CcHsp70-5 | 1917 | 638 | 70.000 | 5.50 | 34.26 | 81.54 | -0.467 | 细胞质Cytoplasm |
| CcHsp70-6 | 1935 | 644 | 70.496 | 5.50 | 38.54 | 82.87 | -0.423 | 细胞质Cytoplasm |
| CcHsp70-7 | 1965 | 654 | 71.593 | 5.56 | 38.44 | 82.22 | -0.445 | 细胞质Cytoplasm |
| CcHsp70-8 | 1980 | 659 | 72.901 | 5.07 | 25.59 | 86.56 | -0.482 | 内质网Endoplasmic reticulum |
| CcHsp90-1 | 2091 | 696 | 78.856 | 8.04 | 40.02 | 88.97 | -0.424 | 线粒体基质 Mitochondrial matrix |
| CcHsp90-2 | 2172 | 723 | 82.812 | 4.97 | 39.90 | 80.24 | -0.681 | 细胞质Cytoplasm |
| CcHsp90-3 | 2331 | 776 | 88.900 | 4.95 | 39.29 | 81.40 | -0.603 | 细胞外Extracellular |

图2
绿豆象HSP超家族基因的系统发育关系、基因编码的蛋白结构和蛋白质保守基序分析 A:31个HSP基因编码的氨基酸序列构建的无根系统发育树,不同颜色区域代表不同CcHsp The unrooted phylogenetic tree was constructed from 31 amino acid sequences encoded by HSP genes, different CcHsps were marked with different colors;B:蛋白质保守基序分析,不同颜色方块代表不同基序Conserved motif of proteins, different color squares represented different motifs;C:绿豆象HSP超家族基因的CDS和UTR分析CDS and UTR analysis of the HSP proteins in C. chinensis"
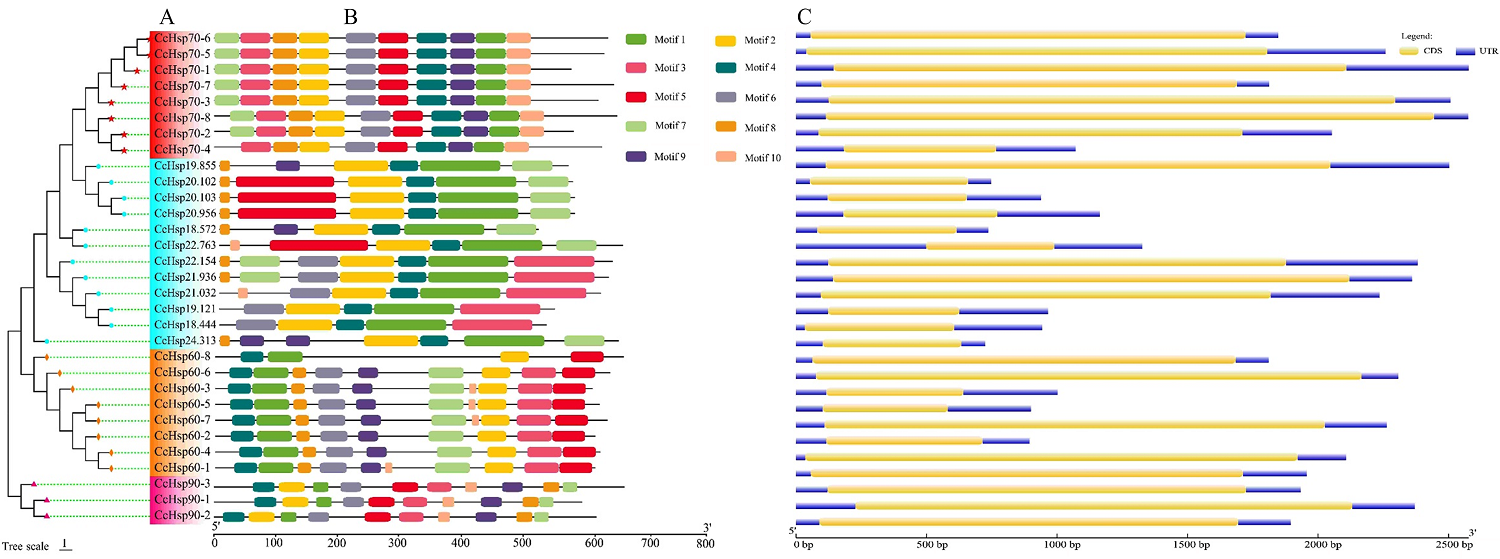
表3
31个CcHsp在绿豆象成虫高、低温转录组中的表达谱"
| 基因ID Gene ID | 基因名 Gene name | 描述 Description | A1 (FPKM) | A2 (FPKM) | A3 (FPKM) | B1 (FPKM) | B2 (FPKM) | B3 (FPKM) | C1 (FPKM) | C2 (FPKM) | C3 (FPKM) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F01_transcript_43299 | CcHsp18.572 | Alpha-crystallin B chain-like | 8.98 | 5.94 | 4.90 | 5188.18 | 3905.53 | 4239.34 | 11.05 | 17.72 | 0 |
| F01_transcript_42933 | CcHsp19.855 | Alpha-crystallin B chain-like | 20.60 | 26.01 | 18.35 | 5155.01 | 5338.97 | 5536.08 | 49.24 | 27.53 | 19.29 |
| F01_transcript_40250 | CcHsp20.102 | Protein lethal (2) essential for life | 8.19 | 12.68 | 11.84 | 14663.45 | 14417.42 | 12560.95 | 12.78 | 42.34 | 10.79 |
| F01_transcript_39920 | CcHsp21.032 | Alpha-crystallin B chain-like | 0 | 0 | 0 | 1.49 | 1.16 | 0.32 | 0 | 0 | 0 |
| F01_transcript_38328 | CcHsp19.121 | Alpha-crystallin B chain-like | 31.83 | 30.27 | 19.40 | 793.80 | 422.20 | 553.81 | 18.71 | 42.25 | 15.67 |
| F01_transcript_28534 | CcHsp18.444 | Alpha-crystallin B chain-like | 0 | 2.17 | 1.50 | 12.66 | 0 | 0 | 0.14 | 0.65 | 0 |
| F01_transcript_27941 | CcHsp20.103 | Protein lethal (2) essential for life | 29.99 | 21.96 | 22.74 | 16.40 | 70.62 | 4.77 | 64.49 | 13.95 | 21.07 |
| F01_transcript_24610 | CcHsp22.154 | Protein lethal (2) essential for life | 26.12 | 27.01 | 16.63 | 1208.61 | 508.81 | 769.67 | 17.83 | 34.94 | 13.28 |
| F01_transcript_24085 | CcHsp20.956 | Protein lethal (2) essential for life | 122.65 | 191.95 | 87.92 | 3007.42 | 3623.59 | 4306.85 | 118.63 | 76.41 | 78.69 |
| F01_transcript_24011 | CcHsp22.763 | Alpha-crystallin B chain-like | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F01_transcript_22315 | CcHsp21.936 | Protein lethal (2) essential for life | 20.37 | 49.74 | 23.53 | 616.41 | 1482.13 | 892.17 | 13.49 | 26.33 | 37.90 |
| F01_transcript_5515 | CcHsp24.313 | Alpha-crystallin B chain-like | 239.53 | 273.50 | 197.18 | 250.14 | 431.63 | 251.55 | 208.14 | 381.78 | 263.16 |
| F01_transcript_128 | CcHsp60-8 | 60 kDa heat shock protein | 152.30 | 368.87 | 363.28 | 445.95 | 297.55 | 250.78 | 237.47 | 397.82 | 120.83 |
| F01_transcript_40995 | CcHsp60-5 | T-complex protein 1 subunit eta-like | 41.21 | 38.76 | 37.33 | 71.18 | 21.85 | 33.86 | 38.20 | 49.72 | 18.06 |
| F01_transcript_20569 | CcHsp60-4 | T-complex protein 1 subunit epsilon | 13.40 | 21.90 | 49.28 | 31.97 | 30.52 | 16.34 | 14.34 | 44.61 | 20.79 |
| F01_transcript_694 | CcHsp60-3 | T-complex protein 1 subunit zeta | 102.08 | 125.65 | 112.61 | 100.87 | 67.07 | 98.74 | 86.41 | 116.36 | 71.93 |
| F01_transcript_639 | CcHsp60-6 | T-complex protein 1 subunit alpha | 46.86 | 86.59 | 94.75 | 95.54 | 54.49 | 94.64 | 71.68 | 122.22 | 69.76 |
| F01_transcript_578 | CcHsp60-2 | T-complex protein 1 subunit beta | 63.48 | 71.65 | 69.67 | 66.85 | 61.50 | 77.62 | 63.93 | 95.76 | 70.86 |
| F01_transcript_501 | CcHsp60-1 | T-complex protein 1 subunit delta | 80.57 | 93.12 | 71.84 | 58.52 | 45.90 | 53.01 | 36.07 | 83.26 | 77.10 |
| F01_transcript_442 | CcHsp60-7 | T-complex protein 1 subunit gamma | 89.12 | 101.69 | 91.83 | 85.56 | 64.15 | 75.93 | 95.03 | 109.45 | 79.94 |
| F01_transcript_73 | CcHsp70-6 | Heat shock protein 70 | 6.03 | 3.74 | 9.44 | 1609.09 | 1163.31 | 1709.61 | 6.05 | 12.11 | 0 |
| F01_transcript_18358 | CcHsp70-2 | Heat shock 70 kDa protein cognate 3 isoform X1 | 426.14 | 211.25 | 179.19 | 536.20 | 53.68 | 35.18 | 105.66 | 82.65 | 21.55 |
| F01_transcript_16333 | CcHsp70-4 | Heat shock 70 kDa protein cognate 3 | 0.31 | 0 | 0 | 0 | 0.77 | 0 | 0 | 0 | 0 |
| F01_transcript_277 | CcHsp70-3 | Heat shock 70 kDa protein cognate 2 | 39.45 | 40.46 | 65.39 | 55.88 | 81.61 | 43.11 | 83.69 | 35.19 | 31.18 |
| F01_transcript_168 | CcHsp70-5 | Heat shock protein 70 | 10.63 | 10.46 | 9.83 | 5992.26 | 4679.61 | 6348.31 | 11.82 | 21.93 | 5.07 |
| F01_transcript_122 | CcHsp70-8 | Heat shock 70 kDa protein cognate 3 isoform X3 | 112.90 | 305.41 | 319.01 | 851.12 | 1148.55 | 644.20 | 274.10 | 555.56 | 385.90 |
| F01_transcript_120 | CcHsp70-1 | Heat shock protein 70 | 0.03 | 1.12 | 2.54 | 133.45 | 574.24 | 386.62 | 1.94 | 1.71 | 1.50 |
| F01_transcript_56 | CcHsp70-7 | Heat shock protein 70 | 0 | 110.53 | 0 | 0 | 2.09 | 0 | 0 | 1.63 | 1.13 |
| F01_transcript_42638 | CcHsp90-1 | Heat shock protein 75 kDa | 20.55 | 22.54 | 21.28 | 21.17 | 22.56 | 23.64 | 18.12 | 22.08 | 30.43 |
| F01_transcript_59 | CcHsp90-3 | Endoplasmin | 254.32 | 274.77 | 261.66 | 361.26 | 319.09 | 296.49 | 136.10 | 214.31 | 168.92 |
| F01_transcript_57 | CcHsp90-2 | Heat shock protein 83 | 1122.57 | 1278.61 | 1487.83 | 3408.15 | 3540.80 | 3795.53 | 758.88 | 1352.60 | 1729.74 |
表4
高、低温胁迫转录组中HSP基因差异表达的统计分析"
| 基因ID Gene ID | A1_Count | A2_Count | A3_Count | B1_Count | B2_Count | B3_Count | 错误发现率 FDR | 差异倍数的 对数值 Log2FC | 基因调节 Gene regulated |
|---|---|---|---|---|---|---|---|---|---|
| F01_transcript_43299 | 83 | 52 | 47 | 45192 | 36760 | 34185 | 1.25E-281 | 8.875292037 | Up |
| F01_transcript_38328 | 326 | 295 | 207 | 7703 | 4431 | 4996 | 6.02E-50 | 4.188931643 | Up |
| F01_transcript_24624 | 63 | 152 | 42 | 22981 | 31416 | 26459 | 1.09E-69 | 6.883143797 | Up |
| F01_transcript_3613 | 125 | 152 | 105 | 23298 | 32452 | 32178 | 3.91E-195 | 7.499200641 | Up |
| F01_transcript_20561 | 0 | 0 | 0 | 54 | 42 | 36 | 6.81E-13 | 3.780936629 | Up |
| F01_transcript_25418 | 23 | 7 | 18 | 19904 | 19924 | 16508 | 1.20E-208 | 9.246980188 | Up |
| F01_transcript_5284 | 33 | 23 | 0 | 3143 | 1904 | 1905 | 0.019906364 | 1.64057389 | Up |
| F01_transcript_36652 | 31 | 52 | 52 | 128 | 278 | 165 | 2.89E-05 | 1.884678396 | Up |
| F01_transcript_43228 | 0 | 0 | 1 | 67 | 149 | 724 | 1.88E-08 | 3.227036338 | Up |
| F01_transcript_33596 | 270 | 269 | 146 | 693 | 709 | 451 | 2.48E-05 | 1.476145857 | Up |
| F01_transcript_41848 | 1258 | 1272 | 1250 | 3286 | 3617 | 2559 | 1.85E-13 | 1.448039781 | Up |
| F01_transcript_28277 | 1 | 3 | 7 | 86 | 296 | 795 | 1.67E-09 | 3.378050236 | Up |
| F01_transcript_6174 | 11 | 6 | 54 | 4940 | 9656 | 5025 | 7.99E-21 | 4.73690697 | Up |
| F01_transcript_18613 | 0 | 12 | 4 | 5653 | 5050 | 3444 | 1.44E-65 | 7.225268629 | Up |
| F01_transcript_17534 | 42 | 21 | 92 | 753 | 630 | 959 | 3.63E-11 | 3.117146973 | Up |
| F01_transcript_42430 | 2164 | 1080 | 1190 | 7186 | 7757 | 5735 | 2.46E-12 | 2.200081315 | Up |
| F01_transcript_38496 | 650 | 797 | 647 | 1878 | 2100 | 1393 | 1.56E-09 | 1.459606834 | Up |
| F01_transcript_40250 | 53 | 77 | 79 | 88274 | 93445 | 68696 | 0 | 9.709356267 | Up |
| F01_transcript_29133 | 1 | 7 | 7 | 1406 | 1481 | 1820 | 1.86E-85 | 1.723134458 | Up |
| F01_transcript_19811 | 32 | 30 | 55 | 154 | 154 | 108 | 3.71E-05 | 1.065158394 | Up |
| F01_transcript_47 | 296002 | 298628 | 475364 | 749443 | 723281 | 659223 | 0.003868698 | 1.109360769 | Up |
| F01_transcript_34439 | 20 | 15 | 20 | 42 | 42 | 36 | 0.039086603 | 1.605846729 | Up |
| F01_transcript_122 | 3620 | 9409 | 10704 | 26414 | 38741 | 19064 | 0.001900569 | 6.502458533 | Up |
| F01_transcript_30785 | 0 | 0 | 0 | 423 | 320 | 442 | 1.43E-52 | 1.382236836 | Up |
| F01_transcript_46898 | 164 | 175 | 56 | 253 | 693 | 263 | 0.02788211 | 4.184196848 | Up |
| F01_transcript_22315 | 242 | 565 | 292 | 6981 | 18172 | 9444 | 2.10E-24 | 4.4489631 | Up |
| F01_transcript_24085 | 1209 | 1799 | 902 | 27990 | 36460 | 37206 | 2.08E-50 | 1.286238774 | Up |
| F01_transcript_25973 | 19 | 16 | 16 | 36 | 65 | 34 | 0.024418377 | 1.601501158 | Up |
| F01_transcript_45548 | 90 | 137 | 174 | 425 | 381 | 428 | 1.92E-05 | 3.316756015 | Up |
| F01_transcript_35814 | 3 | 7 | 3 | 82 | 82 | 60 | 6.56E-14 | 5.119419417 | Up |
| F01_transcript_21658 | 17 | 14 | 17 | 1080 | 557 | 1040 | 5.07E-47 | 3.788445916 | Up |
| F01_transcript_37597 | 601 | 727 | 663 | 11682 | 10180 | 6929 | 1.31E-55 | 1.272772815 | Up |
| F01_transcript_28254 | 31 | 11 | 27 | 77 | 55 | 52 | 0.033458361 | 6.722645203 | Up |
| F01_transcript_73 | 206 | 123 | 339 | 53457 | 42010 | 54200 | 1.57E-75 | 1.4927542 | Up |
| F01_transcript_21952 | 22 | 11 | 52 | 133 | 87 | 98 | 0.021134189 | 1.513921055 | Up |
| F01_transcript_57 | 38595 | 42246 | 53536 | 113486 | 128163 | 120611 | 7.83E-09 | 1.538732717 | Up |
| F01_transcript_23965 | 425 | 382 | 379 | 927 | 1591 | 841 | 2.81E-06 | 7.802804908 | Up |
| F01_transcript_42933 | 223 | 268 | 207 | 52943 | 59323 | 52969 | 0 | 2.20861966 | Up |
| F01_transcript_27028 | 6 | 4 | 3 | 44 | 49 | 18 | 0.000136632 | 1.259693495 | Up |
| F01_transcript_19334 | 197 | 244 | 249 | 536 | 451 | 555 | 1.31E-05 | 1.467376685 | Up |
| F01_transcript_11744 | 43 | 21 | 0 | 2103 | 1367 | 1285 | 0.045002773 | 4.400867923 | Up |
| F01_transcript_27257 | 1970 | 2940 | 2585 | 57913 | 52839 | 57508 | 1.67E-76 | 4.702861053 | Up |
| F01_transcript_24610 | 349 | 344 | 231 | 15406 | 7026 | 9202 | 3.65E-48 | 1.199883723 | Up |
| F01_transcript_33669 | 222 | 189 | 244 | 530 | 461 | 405 | 6.09E-06 | 8.919378418 | Up |
| F01_transcript_168 | 325 | 306 | 314 | 177176 | 150364 | 178888 | 0 | 1.504407815 | Up |
| F01_transcript_120 | 1 | 34 | 86 | 4186 | 19580 | 11567 | 0.036109917 | 6.953691731 | Up |
| F01_transcript_23373 | 17 | 20 | 17 | 5167 | 4047 | 2431 | 3.95E-103 | 1.674610512 | Up |
| F01_transcript_3052 | 452 | 458 | 419 | 1457 | 1384 | 1062 | 1.08E-18 | 5.053735916 | Up |
| F01_transcript_41976 | 16 | 2 | 1 | 920 | 920 | 826 | 1.98E-27 | 3.220154002 | Up |
| F01_transcript_4271 | 0 | 13 | 18 | 434 | 520 | 280 | 4.73E-09 | 1.781266067 | Up |
| F01_transcript_36223 | 8 | 0 | 0 | 24988 | 25210 | 20569 | 0.009242401 | 8.875292037 | Up |
| [1] |
仲建锋, 万正煌, 李莉, 陈宏伟, 伍广洪. 低温和高温对仓储绿豆象的防治效果. 中国农业科学, 2013, 46(1): 54-59. doi: 10.3864/j.issn.0578-1752.2013.01.007.
|
|
|
|
| [2] |
doi: 10.1016/j.ygeno.2020.07.040 |
| [3] |
doi: 10.1007/s12192-011-0286-2 |
| [4] |
doi: 10.1111/ins.2014.21.issue-4 |
| [5] |
doi: 10.1016/j.jinsphys.2009.09.012 pmid: 19782687 |
| [6] |
|
| [7] |
doi: 10.1146/annurev-ento-011613-162107 pmid: 25341107 |
| [8] |
|
| [9] |
doi: 10.1016/j.ygeno.2020.04.019 |
| [10] |
doi: 10.1016/j.cbd.2020.100677 |
| [11] |
doi: 10.1016/j.cbd.2019.03.009 |
| [12] |
doi: 10.1186/s12864-022-08858-1 |
| [13] |
doi: 10.1016/j.ijbiomac.2021.07.186 |
| [14] |
doi: 10.1016/j.ijbiomac.2022.01.014 pmid: 35016971 |
| [15] |
doi: 10.1111/ins.2019.26.issue-1 |
| [16] |
|
| [17] |
doi: 10.1093/nar/gkv1204 |
| [18] |
doi: 10.1093/nar/gkr367 |
| [19] |
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [20] |
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [21] |
doi: 10.1006/meth.2001.1262 |
| [22] |
Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature, 2008, 452(7190): 949-955.
doi: 10.1038/nature06784 |
| [23] |
doi: 10.1186/1471-2229-4-10 |
| [24] |
doi: 10.3390/insects13070570 |
| [25] |
doi: 10.3892/ijmm.2015.2380 |
| [26] |
|
| [27] |
pmid: 8151709 |
| [28] |
doi: 10.1111/1744-7917.12543 pmid: 28980406 |
| [29] |
doi: 10.1016/S2095-3119(19)62808-X |
| [30] |
doi: 10.1007/s12192-021-01198-1 |
| [31] |
doi: 10.3390/insects11120839 |
| [32] |
doi: 10.1016/j.ijbiomac.2021.10.186 |
| [33] |
doi: 10.3390/genes10100775 |
| [34] |
doi: 10.7717/peerj.6992 |
| [1] | 邵红扬, 孟香, 张涛, 陈敏. 响应槲皮素诱导的美国白蛾P450基因及CYP6ZB2功能分析[J]. 中国农业科学, 2023, 56(7): 1322-1332. |
| [2] | 张开京, 何帅帅, 贾利, 胡玉超, 杨德坤, 陆晓民, 张其安, 严从生. 黄瓜DIR家族基因的全基因组鉴定及其表达分析[J]. 中国农业科学, 2023, 56(4): 711-728. |
| [3] | 王壮壮, 董邵云, 周琪, 苗晗, 刘小萍, 徐奎鹏, 顾兴芳, 张圣平. 黄瓜果实维生素C合成关键基因克隆与分析[J]. 中国农业科学, 2023, 56(3): 508-518. |
| [4] | 刘瑞, 赵羽涵, 付忠举, 顾欣怡, 王艳霞, 靳学慧, 杨莹, 吴伟怀, 张亚玲. 黑龙江省和海南省PWL基因家族在稻瘟病菌中的分布及变异[J]. 中国农业科学, 2023, 56(2): 264-274. |
| [5] | 张钰佳, 崔凯文, 段力升, 曹爱萍, 谢全亮, 沈海涛, 王斐, 李鸿彬. 海岛棉CAD和CAD-Like基因家族的鉴定、表达及其对大丽轮枝菌的响应[J]. 中国农业科学, 2023, 56(19): 3759-3771. |
| [6] | 杨惠珍, 杨欢, 吴子璇, 范阔海, 尹伟, 孙盼盼, 钟佳, 孙娜, 李宏全. 猪SsMT-1A和SsMT-2A的原核表达及金属结合特性研究[J]. 中国农业科学, 2023, 56(17): 3461-3478. |
| [7] | 孔乐辉, 宗德乾, 史青尧, 殷盼盼, 巫文玉, 田鹏, 单卫星, 强晓玉. 马铃薯StCYP83基因家族鉴定及其抗晚疫病的功能分析[J]. 中国农业科学, 2023, 56(16): 3124-3139. |
| [8] | 贺丹, 尤啸龙, 何松林, 张明星, 张佼蕊, 华超, 王政, 刘艺平. 芍药胼胝质合成酶基因家族鉴定及PlCalS5功能分析[J]. 中国农业科学, 2023, 56(16): 3183-3198. |
| [9] | 李世佳,吕紫敬,赵锦. 枣R2R3-MYB亚家族基因鉴定及其在果实发育中的表达分析[J]. 中国农业科学, 2022, 55(6): 1199-1212. |
| [10] | 赖春旺, 周小娟, 陈燕, 刘梦雨, 薛晓东, 肖学宸, 林文忠, 赖钟雄, 林玉玲. 龙眼乙烯合成途径基因鉴定及响应ACC处理的分析[J]. 中国农业科学, 2022, 55(3): 558-574. |
| [11] | 郭绍雷, 许建兰, 王晓俊, 宿子文, 张斌斌, 马瑞娟, 俞明亮. 桃XTH家族基因鉴定及其在桃果实贮藏过程中的表达特性[J]. 中国农业科学, 2022, 55(23): 4702-4716. |
| [12] | 陈凤琼, 陈秋森, 林佳昕, 王雅亭, 刘汉林, 梁冰若诗, 邓艺茹, 任春元, 张玉先, 杨凤军, 于高波, 魏金鹏, 王孟雪. 番茄DIR基因家族鉴定及其对非生物胁迫响应的分析[J]. 中国农业科学, 2022, 55(19): 3807-3821. |
| [13] | 郑海霞,高玉林,张方梅,杨超霞,蒋健,朱勋,张云慧,李祥瑞. 马铃薯甲虫热激蛋白基因Ld-hsp70的克隆及温度胁迫下的表达特性[J]. 中国农业科学, 2021, 54(6): 1163-1175. |
| [14] | 郑逢盛,王海华,邬清韬,申权,田建红,彭喜旭,唐新科. 苦荞VQ基因家族的全基因组鉴定及其在叶斑病原与激素处理下的表达谱分析[J]. 中国农业科学, 2021, 54(19): 4048-4060. |
| [15] | 黄金凤,吕天星,王寻,王颖达,王冬梅,闫忠业,刘志. 苹果LRR-RLK基因家族鉴定和表达分析[J]. 中国农业科学, 2021, 54(14): 3097-3112. |
|
||