













中国农业科学 ›› 2023, Vol. 56 ›› Issue (10): 1881-1892.doi: 10.3864/j.issn.0578-1752.2023.10.006
宾羽( ), 张琦, 王春庆, 赵晓春, 宋震(
), 张琦, 王春庆, 赵晓春, 宋震( ), 周常勇(
), 周常勇( )
)
收稿日期:2023-03-02
接受日期:2023-03-08
出版日期:2023-05-16
发布日期:2023-05-17
通信作者:
宋震,E-mail:songzhen@cric.cn。周常勇,E-mail:zhoucy@cric.cn
联系方式:
宾羽,E-mail:bin20196040@swu.edu.cn。
基金资助:
BIN Yu( ), ZHANG Qi, WANG ChunQing, ZHAO XiaoChun, SONG Zhen(
), ZHANG Qi, WANG ChunQing, ZHAO XiaoChun, SONG Zhen( ), ZHOU ChangYong(
), ZHOU ChangYong( )
)
Received:2023-03-02
Accepted:2023-03-08
Published:2023-05-16
Online:2023-05-17
摘要:
【目的】柑橘黄化脉明病毒(citrus yellow vein clearing virus,CYVCV)是威胁我国柑橘产业稳定发展的主要病毒,但其在柑橘上的侵染和致病机制尚不清楚。本文以CYVCV的外壳蛋白(coat protein,CP)为诱饵筛选尤力克柠檬(Citrus limon Burm. f.)cDNA文库,利用生物信息学方法分析与其互作的寄主因子在病毒侵染和致病过程中可能发挥的作用。【方法】Trizol法提取尤力克柠檬叶片总RNA,用SMART法反转录合成First-Strand cDNA,以其为模板通过Long-Distance PCR获得ds cDNA,均一化处理后与线性化pGADT7质粒重组链接构建尤力克柠檬初级cDNA文库,重组质粒转化大肠杆菌DH10B获得尤力克柠檬大肠杆菌cDNA文库并对文库质量进行鉴定;PCR扩增CYVCV的CP序列并构建到载体pGBKT7上,鉴定诱饵质粒对酵母细胞的毒性以及CP蛋白对酵母报告基因的自激活性。将尤力克柠檬cDNA文库质粒转化含有诱饵质粒pGBKT7-CP的Y2H Gold酵母菌株,共转化子依次涂布SD/-Leu-Trp、SD/-Leu-Trp-His/X-α-Gal和SD/-Leu-Trp-His-Ade/X-α-Gal平板,最终筛选蓝色且长势较好的阳性菌落,提取酵母质粒并测序比对获得候选基因,利用Uniprot在线网站的gene ontology(GO)注释候选基因,分析候选互作蛋白的生物学功能。根据分析结果,选取可能参与寄主抗病或症状发展的候选因子,扩增其CDS全长序列并构建到靶标载体pGADT7后分别与pGBKT7-CP共转化酵母细胞进行点对点酵母双杂交互作验证。【结果】尤力克柠檬大肠杆菌cDNA文库滴度为1.02×108 cfu/mL,库容符合试验标准;成功构建酵母双杂交诱饵载体pGBKT7-CP,该载体没有自激活性且对酵母菌没有毒性;在SD/-Leu-Trp-His-Ade/X-α-Gal平板上筛选得到41个酵母阳性克隆,经序列相似性比对,去除重复,共筛得32个候选寄主因子;GO通路注释结果表明这些寄主因子参与多个叶绿体相关的生物过程,包括碳水化合物代谢、光合作用、光刺激反应、代谢分解和生物合成等过程;这32个寄主因子的分子功能多样,包括催化活性、水解酶活性、转移酶活性、蛋白质结合、转录因子活性和翻译因子活性等,其细胞组分涉及叶绿体、类囊体、膜、细胞质、细胞核和高尔基体等。从候选寄主因子中选取14个重要的蛋白与CP的一对一酵母双杂交验证结果表明,CP与这14个蛋白均发生互作。【结论】成功构建了尤力克柠檬cDNA文库,筛选出32个与CYVCV CP互作的尤力克柠檬寄主因子,分析重要的寄主蛋白功能,推测CYVCV CP通过与光系统II放氧强化复合物组分蛋白(PsbP)、光系统I叶绿素结合蛋白(Lhca3)和1,5-二磷酸核酮糖羧化酶小亚基(RbcS)等多个叶绿体相关蛋白的互作,影响光系统稳定、类囊体结构和叶绿素合成,从而导致光合作用降低以及叶绿体形态和功能受损,酵母点对点验证了CP与这些叶绿体相关因子的互作,这为揭示CYVCV CP参与病毒致病的分子机制提供了理论依据。
宾羽, 张琦, 王春庆, 赵晓春, 宋震, 周常勇. 利用酵母双杂交系统筛选与柑橘黄化脉明病毒CP互作的寄主因子[J]. 中国农业科学, 2023, 56(10): 1881-1892.
BIN Yu, ZHANG Qi, WANG ChunQing, ZHAO XiaoChun, SONG Zhen, ZHOU ChangYong. Screening of the Host Factors Interacting with CP of Citrus Yellow Vein Clearing Virus by Yeast Two-Hybrid System[J]. Scientia Agricultura Sinica, 2023, 56(10): 1881-1892.
表1
PCR引物"
| 引物名称Primer name | 引物序列Primer sequence (5′-3′) |
|---|---|
| CP-Nde I-F | TGATCTCAGAGGAGGACCTGCATATGAGCTTCGACTACACTCACCC |
| CP-BamH I-R | TGCGGCCGCTGCAGGTCGACGGATCCTTAGATGTTGAAAGGGGTC |
| pGADT7-F | TAATACGACTCACTATAGGGCGAGCGCCGCCATG |
| pGADT7-R | TGCGGCCGCTGCAGGTCGACGGATCCTTAGATGTTGAAAGGGGTC |
| 1-F | CCCGGGTGGGCATCGATACGGGATCCATGCAAGATGGGACAGTTCG |
| 1-R | TATCTACGATTCATCTGCAGCTCGAGTCAACTAGAGGTTTGGTTAG |
| 2-F | CCCGGGTGGGCATCGATACGGGATCCATGGCGACCATTTCGAAGCTTAC |
| 2-R | TATCTACGATTCATCTGCAGCTCGAGTTACAGACTAACCAGCTTGAG |
| 3-F | CCCGGGTGGGCATCGATACGGGATCCATGACGAAGAATTACCCCAC |
| 3-R | TATCTACGATTCATCTGCAGCTCGAGTTAGGCTTCAGCAAATCCTAG |
| 5-F | ACGACGTACCAGATTACGCTCATATGGCAACCAGACTTATGTTTGAG |
| 5-R | ATCTGCAGCTCGAGCTCGATGGATCCTTAGACATAGCTGTCAATTAAAG |
| 6-F | ACGACGTACCAGATTACGCTCATATGGAAAACTCAAGCCAAGAATC |
| 6-R | ATCTGCAGCTCGAGCTCGATGGATCCTCAAACTTTCAAAAGTTGCATTTTG |
| 9-F | ACGACGTACCAGATTACGCTCATATGGCTAAGTTTGATGATCTTTATC |
| 9-R | ATCTGCAGCTCGAGCTCGATGGATCCCTAGTGGCGGTGGTGCACGC |
| 20-F | ACGACGTACCAGATTACGCTCATATGGCAACCCAAGCTTTGGTTTC |
| 20-R | TATCTACGATTCATCTGCAGCTCGAGTCAGTGAAACTTGAGGCTAG |
| 21-F | ACGACGTACCAGATTACGCTCATATGGATTCTGGGGGTTCAAAGAG |
| 21-R | ATCTGCAGCTCGAGCTCGATGGATCCTTAAGCAGCTGCAACAGTGC |
| 22-F | ACGACGTACCAGATTACGCTCATATGGCTGCTTCACTGAATCTCCC |
| 22-R | ATCTGCAGCTCGAGCTCGATGGATCCTCAACTGCTTTGAAGAAACTTC |
| 23-F | ACGACGTACCAGATTACGCTCATATGAACACGAACGAGCGCAGAGTTG |
| 23-R | ATCTGCAGCTCGAGCTCGATGGATCCTCAATTCTCCTCTGGATGCTTC |
| 24-F | ACGACGTACCAGATTACGCTCATATGAAGAAAGCAGTGTTGAAACTTG |
| 24-R | ATCTGCAGCTCGAGCTCGATGGATCCTCAGCAAATAACACAAGCATTAG |
| 25-F | ACGACGTACCAGATTACGCTCATATGGCTTCCTCAATGATCTCATC |
| 25-R | ATCTGCAGCTCGAGCTCGATGGATCCCTAGACGCCTGGTGGCTTGG |
| 27-F | ACGACGTACCAGATTACGCTCATATGGGTTCCCTTTGCATAAACGAAG |
| 27-R | ATCTGCAGCTCGAGCTCGATGGATCCTCAAAGTAACACATCTGGCTC |
| 29-F | ACGACGTACCAGATTACGCTCATATGGCTTCAACTCAATGCTTCTTG |
| 29-R | ATCTGCAGCTCGAGCTCGATGGATCCTCAAGCAACACTGAAAGAA |
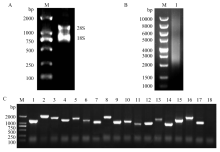
图1
尤力克柠檬cDNA文库构建 A:总RNA琼脂糖凝胶电泳Total RNA of lemon。B:柠檬ds cDNA纯化后的电泳图Electrophoresis of purified ds cDNA of lemon,1:纯化后的ds cDNA Purified ds cDNA。C:cDNA文库插入片段的PCR鉴定PCR identification of inserts in the cDNA library,1—16:随机挑选的16个文库克隆16 clones of cDNA library picked randomly;17:阳性对照Positive control;18:H2O。M:DNA分子标记DNA Marker"
表2
以CYVCV CP为诱饵对柠檬cDNA文库酵母双杂交筛选所获互作候选蛋白"
| 克隆号 Clone number | 登录号 GenBank accession | 蛋白注释ID Uniprot ID | 寄主因子(蛋白名称缩写) Factor of host (Abbreviations of protein name) | 与CP一对一酵母双杂交验证结果 Validation results for one-to-one Y2H with CP |
|---|---|---|---|---|
| 1 | XP_015383675.1 | A0A067E581 | 可能的甲基转移酶20(PMT20)Probable methyltransferase 20 | 互作Interaction |
| 2 | XP_006487560.1 | A0A067ESK0 | 转醛缩酶(TAL)Transaldolase | 互作Interaction |
| 3 | XP_006488195.1 | A0A067G0G4 | L-抗坏血酸过氧化物酶(APX)L-ascorbate peroxidase | 互作Interaction |
| 4 | XP_006494191.1 | V4UB67 | U11/U12小核糖核蛋白35 kDa类蛋白 U11/U12 small nuclear ribonucleoprotein 35 kDa protein-like | — |
| 5 | XP_006479566.1 | V4TUF9 | 类真核翻译起始因子6-2(eIF6) Eukaryotic translation initiation factor 6-2-like | 互作Interaction |
| 6 | NP_001306987.1 | A0A067G915 | WD40重复蛋白(WD40-repeat)WD40 repeat protein | 互作Interaction |
| 7 | XP_006493749.1 | A0A067F2B8 | 琥珀酸-半醛脱氢酶,线粒体亚型X2(SSADH) Succinate-semialdehyde dehydrogenase, mitochondrial isoform X2 | — |
| 8 | XP_006480341.1 | A0A067EMJ8 | 疏水蛋白LTI6B Hydrophobic protein LTI6B | — |
| 9 | XP_006469744.1 | A0A067EA24 | 木葡聚糖内转葡糖基化酶蛋白6(XTH6) Xyloglucan endotransglucosylase/hydrolase protein 6 | 互作Interaction |
| 10 | XP_006466254.1 | A0A067F028 | Ras相关蛋白Rab11A Ras-related protein Rab11A | — |
| 11 | XP_006476755.1 | A0A067FQW5 | 醛脱氢酶家族2成员B4,线粒体(ALDH2B4) Aldehyde dehydrogenase family 2 member B4, mitochondrial | — |
| 12 | XP_024949159.1 | — | 含有RAP结构域的蛋白质,叶绿体 RAP domain-containing protein, chloroplast | — |
| 13 | XP_006481758.1 | A0A067FSV8 | 异质核糖核蛋白Q(HNRNPAQ) Heterogeneous nuclear ribonucleoprotein Q | — |
| 14 | XP_006493717.1 | A0A067F2C0 | 类T复合蛋白1亚基 T-complex protein 1 subunit gamma-like | — |
| 15 | XP_006466732.1 | A0A067GIJ6 | 氨基肽酶P1亚型X1 Aminopeptidase P1 isoform X1 | — |
| 16 | XP_006488080.1 | A0A067GCU3 | 类丝氨酸/苏氨酸蛋白磷酸酶6调节亚基3 Serine/threonine-protein phosphatase 6 regulatory subunit 3-like | — |
| 17 | XP_006473996.1 | A0A067EWB7 | 可能的嘌呤渗透酶5 Probable purine permease 5 | — |
| 18 | NP_001275850.1 | Q8LPM6 | 蔗糖转运蛋白1(SUT)Sucrose transporter 1 | — |
| 19 | XP_006490909.1 | A0A067H1D4 | 转酮醇酶,叶绿体(TK)Transketolase, chloroplast | — |
| 20 | XP_006471243.2 | A0A067DX06 | 光捕获叶绿素a-b结合蛋白3,叶绿体(Lhca3) Light-harvesting chlorophyll a-b binding protein 8, chloroplast | 互作Interaction |
| 21 | XP_006477045.2 | A0A067DAX7 | 类FCS锌指蛋白1(FLZ1)FCS-like zinc finger protein 1 | 互作Interaction |
| 22 | XP_006474598.1 | A0A067GEE4 | 过氧化物还原酶Q,叶绿体(PrxQ)Peroxiredoxin Q, chloroplast | 互作Interaction |
| 23 | XP_006492323.1 | A0A067H7T2 | 通用应激蛋白PHOS34(USP PHOS34)Universal stress protein PHOS34 | 互作Interaction |
| 24 | XP_006490626.1 | A0A067EZZ6 | 重金属相关异戊二烯化植物蛋白39(HIPP39) Heavy metal-associated isoprenylated plant protein 39 | 互作Interaction |
| 25 | XP_006482254.1 | Q84LN2 | 核酮糖二磷酸羧化酶小亚基,叶绿体(RbcS) Ribulose bisphosphate carboxylase (RuBisCO) small subunit, chloroplast | 互作Interaction |
| 26 | XP_006482548.1 | A0A067G267 | V型质子ATP酶亚基d2 V-type proton ATPase subunit d2 | — |
| 27 | XP_006484494.1 | A0A067G359 | F盒/LRR重复蛋白4(FBXL4)F-box/LRR-repeat protein 4 | 互作Interaction |
| 28 | XP_006469469.1 | A0A067D341 | 葡萄糖-6-磷酸1-脱氢酶,类叶绿体(G6PD) Glucose-6-phosphate 1-dehydrogenase, chloroplast-like | — |
| 29 | XP_006485001.1 | A0A067DPL0 | 光系统II放氧强化蛋白2,叶绿体(PsbP) Photosystem II oxygen-evolving enhancer protein 2, chloroplast | 互作Interaction |
| 30 | XP_006476226.1 | — | 未知蛋白Uncharacterized protein | — |
| 31 | XP_006491226.1 | A0A067HEJ5 | 通用植物调节因子1亚型X2 Common plant regulatory factor 1 isoform X2 | — |
| 32 | XP_006478862.1 | A0A067E694 | 类溶质载体家族35成员F2 Solute carrier family 35 member F2-like | — |
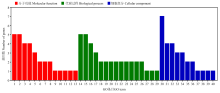
图5
Gene ontology基因功能注释 分子功能Molecular function:1:催化活性Catalytic activity;2:结合Binding;3:水解酶活性Hydrolase activity;4:其他分子功能Other molecular function;5:核苷酸结合Nucleotide binding;6:转运活性Transporter activity;7:蛋白结合Protein binding;8:转移酶活性Transferase activity;9:DNA结合DNA binding;10:DNA结合翻译因子活性DNA-binding transcription factor activity;11:RNA结合RNA binding;12:转录因子活性Translation factor activity;13:酶调节活性Enzyme regulator activity。代谢过程Biological process:14:代谢过程Metabolic process;15:碳水化合物代谢过程Carbohydrate metabolic process;16:其他生物过程Other biological process;17:运输Transport;18:应激响应Response to stress;19:对化学刺激的反应Response to chemical;20:蛋白质代谢过程Protein metabolic process;21:光合作用Photosynthesis;22:前体代谢物和能量的生成Generation of precursor metabolites and energy;23:细胞过程Cellular process;24:细胞成分组织Cellular component organization;25:分解代谢过程Catabolic process;26:生物合成过程Biosynthetic process;27:对光刺激的反应Response to light stimulus;28:含核酶的化合物代谢过程Nucleobase-containing compound metabolic process;29:胞内稳态Cellular homeostasis。细胞组分Cellular component:30:其他细胞组分Other cellular component;31:细胞质Cytoplasm;32:膜Membrane;33:叶绿体Chloroplast;34:类囊体Thylakoid;35:核仁Nucleolus;36:胞浆Cytosol;37:胞外区域Extracellular region;38:细胞核Nucleus;39:内涵体Endosome;40:高尔基体Golgi apparatus"
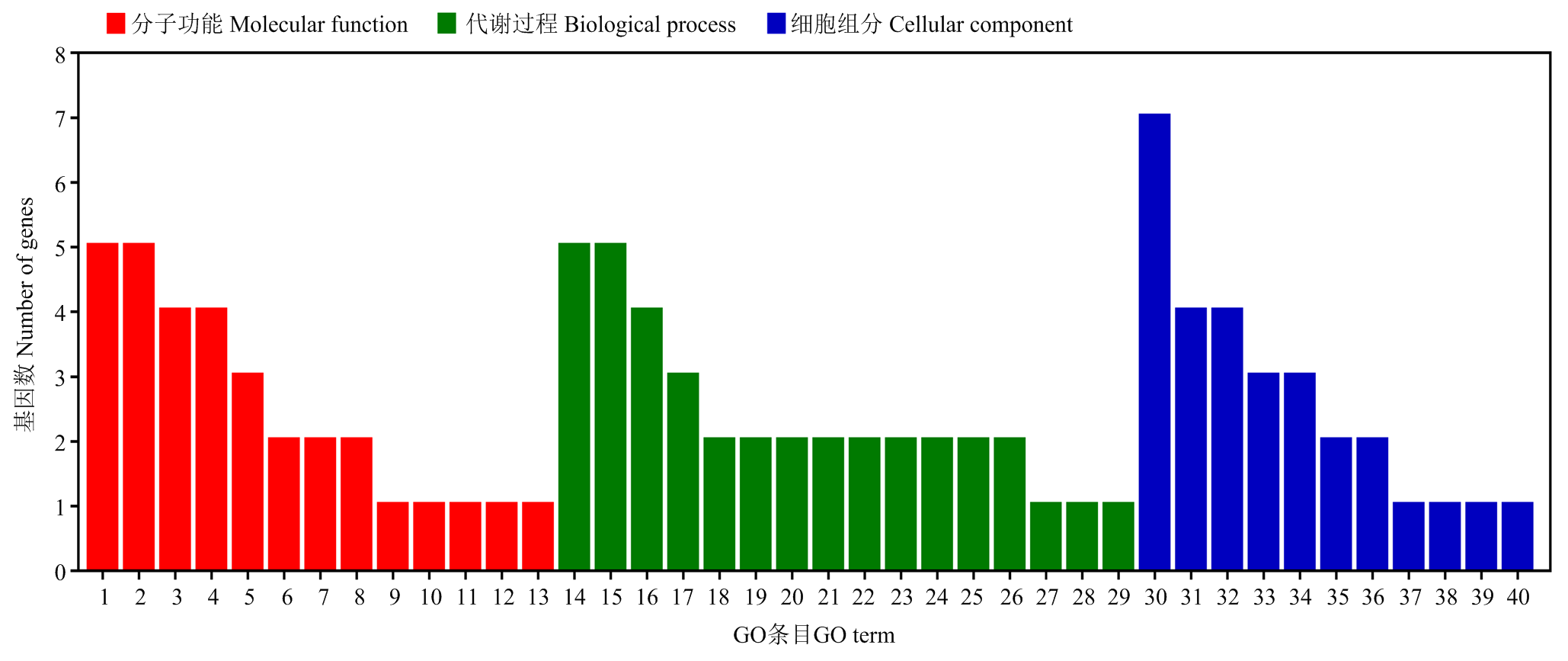

图6
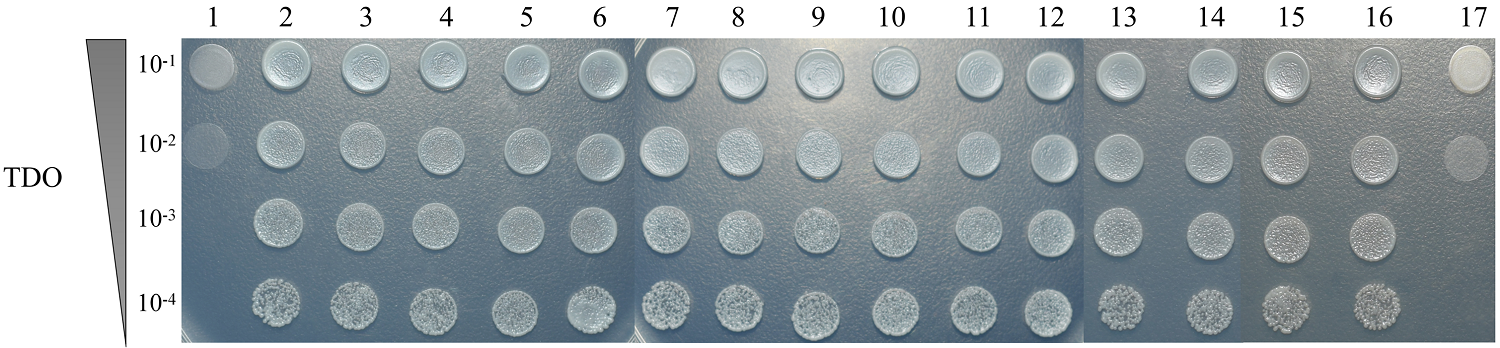
一对一酵母双杂交验证CP与寄主因子的互作 1:pGBKT7-CP+pGADT7;2:pGBKT7-CP+pGADT7-1;3:pGBKT7-CP+pGADT7-2;4:pGBKT7-CP+pGADT7-3;5:pGBKT7-CP+pGADT7-5;6:pGBKT7-CP+pGADT7-6;7:pGBKT7-CP+pGADT7-9;8:pGBKT7-CP+pGADT7-20;9:pGBKT7-CP+pGADT7-21;10:pGBKT7-CP+pGADT7-22;11:pGBKT7-CP+pGADT7-23;12:pGBKT7-CP+pGADT7-24;13:pGBKT7-CP+pGADT7-25;14:pGBKT7-CP+pGADT7-27;15:pGBKT7-CP+pGADT7-29;16:pGBKT7-53+pGADT7-T(正对照Positive control);17:pGBKT7-lam+pGADT7-T(负对照Negative control);TDO:三缺培养基 SD/-Leu-Trp-His medium;10-1—10-4:酵母菌10倍倍比稀释点种Cells of yeast strain Gold were spotted with serial 10-fold dilutions"
| [1] |
doi: 10.1094/PDIS-05-16-0679-RE pmid: 30682317 |
| [2] |
|
| [3] |
doi: 10.3390/v11040329 |
| [4] |
doi: 10.1094/PDIS-02-21-0232-RE |
| [5] |
宾羽, 宋震, 崔甜甜, 周常勇. 柑橘黄化脉明病毒突变体及其构建方法: ZL201910939750.8[P]. (2019-12-31) [2023-03-02].
|
|
|
|
| [6] |
doi: 10.1094/PHYTO-06-12-0140-R |
| [7] |
doi: 10.1007/s00705-015-2423-1 pmid: 25913691 |
| [8] |
doi: 10.1186/s12985-019-1126-8 pmid: 30736799 |
| [9] |
李帅, 蒋西子, 梁伟芳, 陈思涵, 张享享, 左登攀, 胡亚会, 江彤. 利用酵母双杂交系统筛选与草莓镶脉病毒P6蛋白互作的森林草莓寄主因子. 中国农业科学, 2017, 50(18): 3519-3528. doi: 10.3864/j.issn.0578-1752.2017.18.008.
doi: 10.3864/j.issn.0578-1752.2017.18.008 |
|
doi: 10.3864/j.issn.0578-1752.2017.18.008 |
|
| [10] |
pmid: 27757106 |
| [11] |
doi: 10.1094/PHYTO-02-18-0029-R pmid: 29726761 |
| [12] |
doi: 10.1038/s41598-018-19525-5 pmid: 29352213 |
| [13] |
马丹丹. 柑橘黄化脉明病毒在尤力克柠檬中的时空分布及细胞结构变化研究[D]. 重庆: 西南大学, 2016.
|
|
|
|
| [14] |
邓雨青. 柑橘黄化脉明病毒诱导尤力克柠檬细胞程序性死亡研究[D]. 重庆: 西南大学, 2017.
|
|
|
|
| [15] |
doi: 10.1094/MPMI-07-14-0195-R pmid: 25162316 |
| [16] |
doi: 10.1111/mpp.12533 pmid: 28056496 |
| [17] |
doi: 10.1094/MPMI-22-12-1523 pmid: 19888818 |
| [18] |
doi: 10.1007/s00705-013-1778-4 |
| [19] |
doi: 10.3389/fmolb.2018.00024 pmid: 29594130 |
| [20] |
doi: 10.1093/oxfordjournals.pcp.a078472 |
| [21] |
|
| [22] |
doi: 10.1111/nph.2017.214.issue-2 |
| [23] |
doi: 10.1104/pp.112.201459 pmid: 22811433 |
| [24] |
doi: 10.1007/s11262-011-0596-6 pmid: 21400205 |
| [25] |
doi: 10.1104/pp.112.209213 |
| [26] |
|
| [27] |
pmid: 16244145 |
| [28] |
doi: 10.1007/s11120-008-9359-1 |
| [29] |
doi: 10.1016/j.bbabio.2009.03.004 |
| [30] |
doi: 10.1016/j.plaphy.2014.01.001 pmid: 24477118 |
| [31] |
doi: 10.3389/fpls.2016.00084 pmid: 26904056 |
| [32] |
doi: 10.1094/MPMI-02-14-0035-R pmid: 24940990 |
| [33] |
doi: 10.1093/mp/sst158 pmid: 24214893 |
| [34] |
doi: 10.1186/s12864-023-09151-5 |
| [1] | 李雨泽,朱嘉伟,林蔚,蓝茉莹,夏黎明,张艺粒,罗聪,黄桂香,何新华. 香水柠檬RHF2A的克隆与互作蛋白的筛选[J]. 中国农业科学, 2022, 55(24): 4912-4926. |
| [2] | 李天聪,朱行,魏宁,龙凤,武建颖,张燕,董金皋,申珅,郝志敏. 玉米大斑病菌SC35同源基因表达规律与互作分析[J]. 中国农业科学, 2021, 54(4): 733-743. |
| [3] | 马琳,温红雨,王学敏,高洪文,庞永珍. 紫花苜蓿MsMAX2的克隆及功能研究[J]. 中国农业科学, 2021, 54(19): 4061-4069. |
| [4] | 孙雨晨,贾瑞璞,范阔海,孙娜,孙耀贵,孙盼盼,李宏全,尹伟. 猪I型补体受体与C3b活性片段相互结合的体外检测[J]. 中国农业科学, 2021, 54(19): 4243-4254. |
| [5] | 马志敏,许建建,段玉,王春庆,苏越,张琦,宾羽,周常勇,宋震. 柑橘黄化脉明病毒RT-RPA检测方法的建立[J]. 中国农业科学, 2021, 54(15): 3241-3249. |
| [6] | 杨宏凯,杨晶文,沈建国,蔡伟,高芳銮. 辣椒脉斑驳病毒的多基因联合检测与鉴定[J]. 中国农业科学, 2020, 53(16): 3412-3420. |
| [7] | 许海峰,杨官显,张静,邹琦,王意程,曲常志,姜生辉,王楠,陈学森. 苹果MdWRKY18和MdWRKY40参与盐胁迫途径分子机理研究[J]. 中国农业科学, 2018, 51(23): 4514-4521. |
| [8] | 王玉奎,白晓璟,廉小平,张贺翠,罗绍兰,蒲敏,左同鸿,刘倩莹,朱利泉. 甘蓝BoSPx的克隆与表达分析[J]. 中国农业科学, 2018, 51(22): 4328-4338. |
| [9] | 赵青青,李俊平,梁立滨,黄山雨,周陈陈,赵玉辉,王倩,周圆,姜丽,陈化兰,李呈军. 流感病毒PA蛋白与宿主蛋白PCBP1的相互作用[J]. 中国农业科学, 2018, 51(17): 3389-3396. |
| [10] | 李帅,蒋西子,梁伟芳,陈思涵,张享享,左登攀,胡亚会,江彤 . 利用酵母双杂交系统筛选与草莓镶脉病毒P6蛋白互作的森林草莓寄主因子[J]. 中国农业科学, 2017, 50(18): 3519-3528. |
| [11] | 茹京娜,于太飞,陈隽,陈明,周永斌,马有志,徐兆师,闵东红. 小麦锌指转录因子TaDi19A对低温的响应及其互作蛋白的筛选[J]. 中国农业科学, 2017, 50(13): 2411-2422. |
| [12] | 赵娟莹,刘佳明,冯志娟,陈明,周永斌,陈隽,徐兆师,郭长虹. 大豆锌指转录因子GmDi19-5对高温的响应及互作蛋白的筛选[J]. 中国农业科学, 2017, 50(12): 2389-2398. |
| [13] | 袁敏,邢继红,王莉,葛伟娜,郭棣,张岚. 拟南芥开花抑制因子TFL1与GRFs蛋白的相互作用[J]. 中国农业科学, 2017, 50(10): 1772-1780. |
| [14] | 王宇秋,李国邦,杨娟,黎良,赵志学,樊晶,王文明. 稻曲病菌侵染水稻颖花的酵母双杂交cDNA文库构建与应用[J]. 中国农业科学, 2016, 49(5): 865-873. |
| [15] | 张贺翠,柳菁,廉小平,曾静,杨昆,张学杰,杨丹,施松梅,高启国,朱利泉. 甘蓝ROH1与EXO70A1的表达与相互作用[J]. 中国农业科学, 2016, 49(4): 775-783. |
|
||