













中国农业科学 ›› 2022, Vol. 55 ›› Issue (6): 1047-1063.doi: 10.3864/j.issn.0578-1752.2022.06.001
宋松泉1,2( ),刘军1(
),刘军1( ),唐翠芳3,程红焱2,王伟青2,张琪1,张文虎1,高家东1
),唐翠芳3,程红焱2,王伟青2,张琪1,张文虎1,高家东1
收稿日期:2021-08-12
接受日期:2021-10-08
出版日期:2022-03-16
发布日期:2022-03-25
通讯作者:
宋松泉,刘军
基金资助:
SONG SongQuan1,2( ),LIU Jun1(
),LIU Jun1( ),TANG CuiFang3,CHENG HongYan2,WANG WeiQing2,ZHANG Qi1,ZHANG WenHu1,GAO JiaDong1
),TANG CuiFang3,CHENG HongYan2,WANG WeiQing2,ZHANG Qi1,ZHANG WenHu1,GAO JiaDong1
Received:2021-08-12
Accepted:2021-10-08
Online:2022-03-16
Published:2022-03-25
Contact:
SongQuan SONG,Jun LIU
摘要:
耐脱水性是指生物体或组织在丧失所有或几乎所有细胞水分的状态下而不产生不可逆损伤的存活能力。种子的耐脱水性是植物在长期进化过程中保证物种生存和繁衍的适应性机制,在植物种子(质)资源保存中起关键作用。种子的耐脱水性是一个复杂的性状,其分子机理至今尚不清楚。为此,本文综述了种子耐脱水性的生理及分子机制的研究进展。研究发现,正常性种子的耐脱水性是在发育过程中逐渐形成的,在生理成熟期达到峰值;顽拗性种子在整个发育过程中对脱水敏感,不具有成熟脱水的发育阶段。成熟的正常性种子在吸胀初期保持对重新脱水的耐性,随着萌发进程,种子的耐脱水性逐渐下降,最后完全丧失;在萌发初期,种子的耐脱水性可以重建,不同组织具有不同的耐脱水性。种子和胚的耐脱水性程度与其线粒体的呼吸活性下降呈负相关性,顽拗性种子的呼吸活性高于正常性种子。脱水过程中,耐脱水性胚(轴)的H2O2含量、超氧阴离子自由基(·O2-)的产生速率和硫代巴比妥酸活性产物的含量显著低于脱水敏感性胚(轴),而活性氧清除(包括酶促和非酶促)系统的活性明显高于脱水敏感性胚(轴)。种子成熟过程中,胚胎发育晚期丰富(LEA)蛋白、小分子量热休克蛋白和非还原性棉子糖家族寡聚糖的积累与耐脱水性的形成密切相关。B3转录因子的AFL亚家族(包括ABI3(ABA INSENSITIVE 3)、FUS3(FUSCA3)和LEC2(LEAFY COTYLEDON 2))通过正向调控贮藏物和保护性蛋白的积累增加种子(胚)的耐脱水性。在整个种子发育过程中,DNA甲基化水平显著增加,随后在种子萌发过程中逐渐降低;与发育早期阶段的胚和幼苗相比,成熟胚具有较高水平的基因组甲基化。在种子中,平行的ABA和DOG1(DELAY OF GERMINATION 1)信号转导途径激活棉子糖家族寡聚糖的合成、LEA基因和HSP基因的表达,从而调控耐脱水性的起始和向休眠转变。最后,本文提出了该领域需要进一步研究的科学问题,包括利用种子及其组织的不同耐脱水性重建其模式研究系统;种子的萌发能力、耐脱水性和休眠特性都是在发育过程中起始和完成的,它们之间的相互关系仍不清楚;种子中同时存在核心ABA信号途径和DOG1信号途径,这两条途径在ABI3或者ABI3下游汇合,在种子脱水过程中哪条途径优先响应?又是如何协调?本文将为全面理解种子耐脱水性的生理及其分子机制、提高农作物的胁迫抗性与产量、改善资源库的贮藏条件和长期保存植物种子(质)资源提供参考。
宋松泉,刘军,唐翠芳,程红焱,王伟青,张琪,张文虎,高家东. 种子耐脱水性的生理及分子机制研究进展[J]. 中国农业科学, 2022, 55(6): 1047-1063.
SONG SongQuan,LIU Jun,TANG CuiFang,CHENG HongYan,WANG WeiQing,ZHANG Qi,ZHANG WenHu,GAO JiaDong. Research Progress on the Physiology and Its Molecular Mechanism of Seed Desiccation Tolerance[J]. Scientia Agricultura Sinica, 2022, 55(6): 1047-1063.
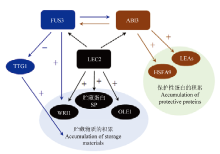
图1
B3转录因子在种子成熟过程中的作用[11] FUS3抑制TTG1(TRANSPARENT TESTA GLABRA 1)转录因子,一个与脂肪酸和贮藏蛋白生物合成相关基因的负调控因子;以及正调控脂肪酸生物合成诱导因子WRI1(WRINKLED 1);因此,FUS3间接地正向影响贮藏物的积累。FUS3也在子叶的两侧调控ABI3表达。LEC2调控其他B3转录因子FUS3和ABI3,阻止花青素和叶绿素的积累,以及通过正调控WRI1和OLE1参与增加脂肪酸的生物合成和贮藏;LEC2也正调控2S和12S贮藏蛋白的表达。ABI3调控胚轴和子叶中FUS3的表达,以及通过正调控HSFA9转录因子间接参与热休克保护性蛋白的积累;ABI3是胚胎发生晚期丰富(LEA)保护性蛋白的主要调控因子"
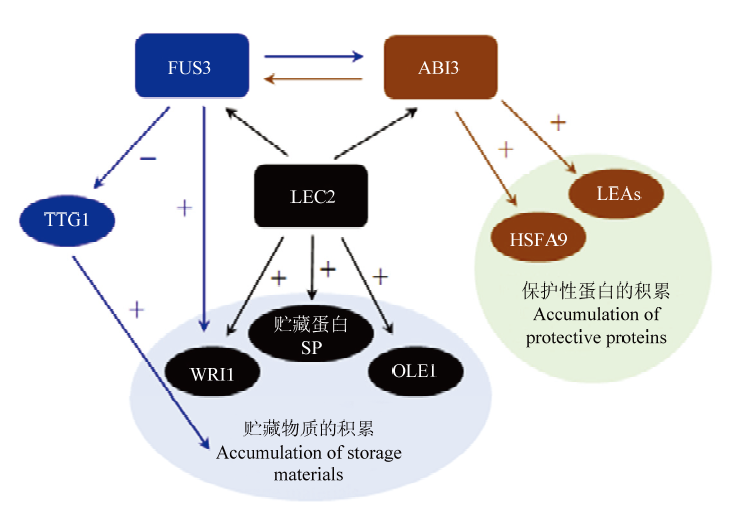
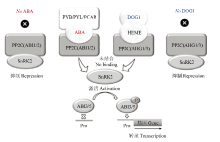
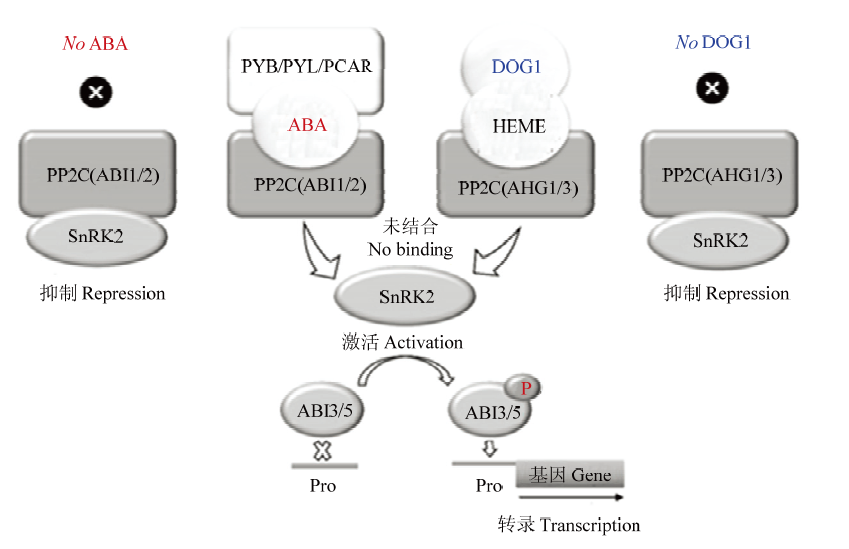
图2
种子中脱落酸(ABA)和DOG1(DELAY OF GERMINATION 1)信号转导途径[12] DOG1信号转导途径的关键组分是血红素分子和由AHG1和AHG3编码的PP2C。PCAR-ABA-PP2C和/或DOG1-血红素-PP2C的三重复合物阻断PP2C与SnRK2的结合。活化的SnRK2使ABI3和ABI5磷酸化,ABI3和ABI5与ABA控制的基因的启动子(Pro)结合。在种子中,平行的ABA和DOG1信号转导途径激活棉子糖家族寡聚糖(RFO)的合成、LEA和HSP的表达,从而调控耐脱水性的起始和向休眠转变。PYB/PYL/PCAR,pyrabactin resistance (PYR)/PYR-like/regulatory component of abscisic acid receptor;AHG,ABA过敏感萌发;PP2C,A组2C类蛋白磷酸酶;SnRK2,亚类Ⅲ蔗糖非发酵-1-相关蛋白激酶2"
| [1] | BEWLEY J D, BRADFORD K J, HILHORST H W M, NONOGAKI H. Seeds: Physiology of Development, Germination and Dormancy. 3rd ed. New York: Springer, 2013. |
| [2] | BLACK M, BEWLEY J D, HALMER P. The Encyclopedia of Seed. Science, Technology and Uses. Oxfordshire: CAB International, 2006. |
| [3] |
LEPRINCE O, BUITINK J. Desiccation tolerance: From genomics to the field. Plant Science, 2010,179:554-564.
doi: 10.1016/j.plantsci.2010.02.011 |
| [4] |
OLIVER M J, FARRAN T J M, HILHORST H W M, MUNDREE S, WILLIAMS B, BEWLEY J D. Desiccation tolerance: Avoiding cellular damage during drying and rehydration. Annual Review of Plant Biology, 2020,71:435-460.
doi: 10.1146/annurev-arplant-071219-105542 |
| [5] | SONG S Q, BERJAK P, PAMMENTER N. Desiccation sensitivity of Trichilia dregeana Sond. axes and antioxidant role of ascorbic acid. Acta Botanica Sinica, 2004,46:803-810. |
| [6] | 傅家瑞, 宋松泉. 顽拗性种子生物学. 北京: 中国科学文化出版社, 2004. |
| FU J R, SONG S Q. Recalcitrant Seed Biology. Beijing: China Science and Culture Press, 2004. (in Chinese) | |
| [7] |
PAMMENTER N W, BERJAK P. Physiology of desiccation-sensitive (recalcitrant) seeds and the implications for cryopreservation. International Journal of Plant Science, 2014,175:21-28.
doi: 10.1086/673302 |
| [8] |
KAN J, SONG S Q. Effects of dehydration, chilling, light, phytohormones and nitric oxide on germination of Pistia stratiotes seeds. Seed Science and Technology, 2008,36:38-45.
doi: 10.15258/sst |
| [9] | BERJAK P, PAMMENTER N W. Implications of the lack of desiccation tolerance in recalcitrant seeds. Frontiers in Plant Science, 2013,4:478. |
| [10] | BERJAK P, PAMMENTER N W. Recalcitrant seeds//BENECH- ARNOLD R L, SNCHEZ R A, eds. Handbook of Seed Physiology: Applications to Agriculture. New York: Haworth Press, 2014: 305-345. |
| [11] |
KIJAK H, RATAJCZAK E. What do we know about the genetic basis of seed desiccation tolerance and longevity? International Journal of Molecular Science, 2020,21:3612.
doi: 10.3390/ijms21103612 |
| [12] |
SMOLIKOVA G, LEONOVA T, VASHURINA N, FROLOV A, MEDVEDEV S. Desiccation tolerance as the basis of long-term seed viability. International Journal of Molecular Sciences, 2021,22:101.
doi: 10.3390/ijms22010101 |
| [13] |
WANG W Q, WANG Y, SONG X J, ZHANG Q, CHENG H Y, LIU J, SONG S Q. Proteomic analysis of desiccation tolerance and its re-establishment in different embryo axis tissues of germinated pea seeds. Journal of Proteome Research, 2021,20:2352-2363.
doi: 10.1021/acs.jproteome.0c00860 |
| [14] |
XU X, LEGAY S, SERGEANT K, ZORZAN S, LECLERCQ C C, CHARTON S, GIAROLA V, LIU X, CHALLABATHULA D, RENAUT J, HAUSMAN J F, BARTELS D, GUERRIERO E. Molecular insights into plant desiccation tolerance: Transcriptomics, proteomics and targeted metabolite profiling in Craterostigma plantagineum. The Plant Journal, 2021,107:377-398.
doi: 10.1111/tpj.v107.2 |
| [15] |
WU J H, WANG W Q, SONG S Q, CHENG H Y. Reactive oxygen species scavenging enzymes and down-adjustment of metabolism level in mitochondria associated with desiccation-tolerance acquisition of maize embryo. Journal of Integrative Plant Biology, 2009,51:638-645.
doi: 10.1111/jipb.2009.51.issue-7 |
| [16] |
WANG W Q, YE J Q, ROGOWSKA-WRZESINSKA A, WOJDYLA K, JENSEN O N, MØLLER I M, SONG S Q. Proteomic comparison between maturation drying and prematurely imposed drying of Zea mayz seeds reveals a potential role of maturation drying in preparing proteins for seed germination, seedling vigor, and pathogen resistance. Journal of Proteome Research, 2013,13:606-626.
doi: 10.1021/pr4007574 |
| [17] |
HUANG H, MØLLER I M, SONG S Q. Proteomics of desiccation tolerance during development and germination of maize embryos. Journal of Proteomics, 2012,75:1247-1262.
doi: 10.1016/j.jprot.2011.10.036 |
| [18] |
HUANG H, SONG S Q, WU X J. Response of Chinese wampee axes and maize embryos on dehydration at different rates. Journal of Integrative Plant Biology, 2009,51:67-74.
doi: 10.1111/jipb.2008.51.issue-1 |
| [19] |
宋松泉, BERJAK P, PAMMENTER N W. Temporal pattern of changes in desiccation tolerance during imbibition of Pisum sativum seeds. 云南植物研究, 2009,31:239-246.
doi: 10.3724/SP.J.1143.2009.08216 |
|
SONG S Q, BERJAK P, PAMMENTER N W. Temporal pattern of changes in desiccation tolerance during imbibition of Pisum sativum seeds. Acta Botanica Yunnanica, 2009,31:239-246. (in Chinese)
doi: 10.3724/SP.J.1143.2009.08216 |
|
| [20] |
WANG W Q, CHENG H Y, MØLLER I M, SONG S Q. The role of recovery of mitochondrial structure and function in desiccation tolerance of pea seeds. Physiologia Plantarum, 2012,144:20-34.
doi: 10.1111/ppl.2012.144.issue-1 |
| [21] | SONG S Q, FU J R. Studies on desiccation sensitivity and peroxidation of membrane lipids in lychee (Litch chinensis Sonn.) seeds. Chinese Science Bulletin, 1992,37:1470-1473. |
| [22] |
CHENG H Y, SONG S Q. Possible involvement of reactive oxygen species scavenging enzymes in desiccation sensitivity of Antiaris toxicaria seeds and axes. Journal of Integrative Plant Biology, 2008,50:1549-1556.
doi: 10.1111/jipb.2008.50.issue-12 |
| [23] | 宋松泉, 傅家瑞. 黄皮种子脱水敏感性与脂质过氧化作用. 植物生理学报, 1997,25:163-168. |
| SONG S Q, FU J R. Desiccation-sensitivity and lipid peroxidation in Chinese wampee [Clausena lansium (Lour.) Skeels] seeds. Acta Phytophysiologica Sinica, 1997,25:163-168. (in Chinese) | |
| [24] |
王伟青, 程红焱, 刘树君, 宋松泉. 黄皮种子线粒体呼吸速率和活性氧清除酶对脱水的响应及其生态学意义. 植物生态学报, 2012,36:870-879.
doi: 10.3724/SP.J.1258.2012.00870 |
|
WANG W Q, CHENG H Y, LIU S J, SONG S Q. Response of respiratory rate and reactive oxygen species scavenging enzyme activity in seed mitochondria of Clausena lansium dehydration and its ecological significance. Chinese Journal Plant Ecology, 2012,36:870-879. (in Chinese)
doi: 10.3724/SP.J.1258.2012.00870 |
|
| [25] |
OBROUCHEVA N V, SINKEVICH I A, LITYAGINA S V. Physiological aspects of seed recalcitrance: A case study on the tree Aesculus hippocastanum. Tree Physiology, 2016,36:1127-1150.
doi: 10.1093/treephys/tpw037 |
| [26] |
SONG S Q, TIAN M H, KAN J, CHENG H Y. The response difference of mitochondria in recalcitrant Antiaris toxicaria axes and orthodox Zea mays embryos to dehydration injury. Journal of Integrative Plant Biology, 2009,51:646-653.
doi: 10.1111/jipb.2009.51.issue-7 |
| [27] |
STAVRINIDES A K, DUSSERT S, COMBES M C, FOCK-BASTIDE I, SEVERAC D, MINIER J, BASTOS-SIQUEIRA A, DEMOLOMBE V, HEM S, LASHERMESW P, JOËT T. Seed comparative genomics in three coffee species identify desiccation tolerance mechanisms in intermediate seeds. Journal of Experimental Botany, 2020,71:1418-1433.
doi: 10.1093/jxb/erz508 |
| [28] |
LEPRINCE O, BUITINK J, HOEKSTRA F A. Axes and cotyledons of recalcitrant seeds of Castanea sativa Mill. exhibit contrasting responses of respiration to drying in relation to desiccation sensitivity. Journal of Experimental Botany, 1999,50:1515-1524.
doi: 10.1093/jxb/50.338.1515 |
| [29] |
MØLLER I M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annual Review of Plant Physiology and Plant Molecular Biology, 2001,52:561-591.
doi: 10.1146/arplant.2001.52.issue-1 |
| [30] |
BAILLY C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochemical Journal, 2019,476:3019-3032.
doi: 10.1042/BCJ20190159 |
| [31] |
DEL RÍO L A. ROS and RNS in plant physiology: An overview. Journal of Experimental Botany, 2015,66:2827-2837.
doi: 10.1093/jxb/erv099 |
| [32] |
MITTLER R. ROS are good. Trends in Plant Science, 2017,22:11-19.
doi: 10.1016/j.tplants.2016.08.002 |
| [33] | DEMIDCHIK V. Reactive oxygen species and their role in plant oxidative stress//SHABALA S, ed. Plant Stress Physiology. CABI: Wallingford, 2017. |
| [34] |
MULLINEAUX P M, BAKER N R. Oxidative stress: Antagonistic signaling for acclimation or cell death? Plant Physiology, 2010,154:521-525.
doi: 10.1104/pp.110.161406 |
| [35] |
JEEVAN KUMAR S P, RAJENDRA P S, BANERJEE R, THAMMINENI C. Seed birth to death: Dual functions of reactive oxygen species in seed physiology. Annals of Botany-London, 2015,116:663-668.
doi: 10.1093/aob/mcv098 |
| [36] |
SANO N, RAJJOU L, NORTH H M, DEBEAUJON I, MARION-POLL A, SEO M. Staying alive: Molecular aspects of seed longevity. Plant Cell and Physiology, 2016,57:660-674.
doi: 10.1093/pcp/pcv186 |
| [37] | COLVILLE L, KRANNER I. Desiccation tolerant plants as model systems to study redox regulation of protein thiols. Plant Growth Regulation, 2016,2:241-255. |
| [38] |
FOYER C H, NOCTOR G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. The Plant Cell, 2005,17:1866-1875.
doi: 10.1105/tpc.105.033589 |
| [39] |
RATAJCZAK E, MAŁECKA A, CIERESZKO I, STASZAK A M. Mitochondria are important determinants of the aging of seeds. International Journal of Molecular Science, 2019,20:1568.
doi: 10.3390/ijms20071568 |
| [40] |
CHEN D, LI Y, FANG T, SHI X, CHEN X. Specific roles of tocopherols and tocotrienols in seed longevity and germination tolerance to abiotic stress in transgenic rice. Plant Science, 2016,244:31-39.
doi: 10.1016/j.plantsci.2015.12.005 |
| [41] |
KUREK K, PLITTA-MICHALAK B, RATAJCZAK E. Reactive oxygen species as potential drivers of the seed aging process. Plants, 2019,8:174.
doi: 10.3390/plants8060174 |
| [42] |
KRANNER I, MINIBAYEVA F V, BECKETT R P, SEAL C E. What is stress? Concepts, definitions and applications in seed science. New Phytologist, 2010,188:655-673.
doi: 10.1111/nph.2010.188.issue-3 |
| [43] |
ROACH T, NAGEL M, BÖRNER A, EBERLE C, KRANNER I. Changes in tocochromanol and glutathione reveal differences in the mechanisms of seed ageing under seed bank conditions and controlled deterioration in barley. Environmental and Experimental Botany, 2018,156:8-15.
doi: 10.1016/j.envexpbot.2018.08.027 |
| [44] |
SHVACHKO N A, KHLESTKINA E K. Molecular genetic bases of seed resistance to oxidative stress during storage. Vavilov Journal Genetics and Breeding, 2020,24:451-458.
doi: 10.18699/VJ20.637 |
| [45] | LEPRINCE O, PELLIZZARO A, BERRIRI S, BUITINK J. Late seed maturation: Drying without dying. Journal of Experimental Botany, 2017,68:827-841. |
| [46] |
MARQUES A, BUIJS G, LIGTERINK W, HILHORST H. Evolutionary ecophysiology of seed desiccation sensitivity. Functional Plant Biology, 2018,45:1083.
doi: 10.1071/FP18022 |
| [47] |
DURE L I I I, GALAU G A. Developmental biochemistry of cottonseed embryogenesis and germination: XIII. Regulation of the biosynthesis of the principal storage proteins. Plant Physiology, 1981,68:187-194.
doi: 10.1104/pp.68.1.187 |
| [48] | BATTAGLIA M, COVARRUBIAS A A. Late embryogenesis abundant (LEA) proteins in legumes. Frontiers in Plant Science, 2013,4:190. |
| [49] |
COSTA M C D, COOPER K, HILHORST H W M, FARRANT J M. Orthodox seeds and resurrection plants: Two of a kind? Plant Physiology, 2017,175:589-599.
doi: 10.1104/pp.17.00760 |
| [50] |
AMARA I, ZAIDI I, MASMOUDI K, LUDEVID M D, PAGÈS M, GODAY A, BRINI F. Insights into late embryogenesis abundant (LEA) proteins in plants: From structure to the functions. American Journal of Plant Sciences, 2014,5:3440-3455.
doi: 10.4236/ajps.2014.522360 |
| [51] |
CANDAT A, PASZKIEWICZ G, NEVEU M, GAUTIER R, LOGAN D C, AVELANGE-MACHEREL M H, MACHEREL D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. The Plant Cell, 2014,26:3148-3166.
doi: 10.1105/tpc.114.127316 |
| [52] |
HUNDERTMARK M, HINCHA D K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics, 2008,9:118.
doi: 10.1186/1471-2164-9-118 |
| [53] |
ARTUR M A S, ZHAO T, LIGTERINK W, SCHRANZ E, HILHORST H W M. Dissecting the genomic diversification of late embryogenesis abundant (LEA) protein gene families in plants. Genome Biology and Evolution, 2019,11:459-471.
doi: 10.1093/gbe/evy248 |
| [54] |
CHEN C, ZABAD S, LIU H, WANG W, JEFFERY C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Research, 2018,46:D640-D644.
doi: 10.1093/nar/gkx1043 |
| [55] |
JIN X, CAO D, WANG Z, MA L, TIAN K, LIU Y, GONG Z, ZHU X, JIANG C, LI Y. Genome-wide identification and expression analyses of the LEA protein gene family in tea plant reveal their involvement in seed development and abiotic stress responses. Scientific Reports, 2019,9:14123.
doi: 10.1038/s41598-019-50645-8 |
| [56] |
OLVERA-CARRILLO Y, CAMPOS F, REYES J L, GARCIARRUBIO A, COVARRUBIAS A A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiology, 2010,154:373-390.
doi: 10.1104/pp.110.158964 |
| [57] |
FARRANT J M, PAMMENTER N W, BERJAK P. Seed development in relation to desiccation tolerance: A comparison between desiccation-sensitive (recalcitrant) seeds of Avicennia marina and desiccation-tolerant types. Seed Science Research, 1993,3:1-13.
doi: 10.1017/S0960258500001513 |
| [58] |
DELAHAIE J, HUNDERTMARK M, BOVE J, LEPRINCE O, ROGNIAUX H, BUITINK J. LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. Journal of Experimental Botany, 2013,64:4559-4573.
doi: 10.1093/jxb/ert274 |
| [59] | JIN X, LIU D, MA L, GONG Z, CAO D, LIU Y, LI Y, JIANG C. Transcriptome and expression profiling analysis of recalcitrant tea (Camellia sinensis L.) seeds sensitive to dehydration. International Journal of Genomics, 2018,2018:5963797. |
| [60] |
DUSSERT S, SERRET J, BASTOS-SIQUEIRA A, MORCILLO F, DÈCHAMP E, ROFIDAL V, LASHERMES P, ETIENNE H, JOËT T. Integrative analysis of the late maturation programme and desiccation tolerance mechanisms in intermediate coffee seeds. Journal of Experimental Botany, 2018,69:1583-1597.
doi: 10.1093/jxb/erx492 |
| [61] | KALEMBA E M, PUKACKA S. Possible role of LEA proteins and sHSPs in seed protection: A short review. Biology Letters, 2007,44:3-16. |
| [62] | KAUR H, PETLA B P, KAMBLE N U, SINGH A, RAO V, SALVI P, GHOSH S, MAJEE M. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Frontiers in Plant Science, 2015,6:713. |
| [63] |
NOVER L, BHARTI K, DÖRING P, MISHRA S K, GANGULI A, SCHARF K D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress and Chaperones, 2001,6:177.
doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2 |
| [64] |
NETO V G, BARBOSA R R, CAROSIO M G A, FERREIRA A G, FERNANDEZ L G, DE CASTRO R D, LIGTERINK W, HILHORST H, RIBERIRO P R. Sequence analysis of Ricinus communis small heat shock protein (sHSP) subfamily and its role in abiotic stress responses. Industrial Crops and Products, 2020,152:112541.
doi: 10.1016/j.indcrop.2020.112541 |
| [65] |
DEKKERS B J W, HE H, HANSON J, WILLEMS L A J, JAMER D C L, CUEFF G, RAJJOU L, HILHORST H W M, BENTSINK L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. The Plant Journal, 2016,85:451-465.
doi: 10.1111/tpj.13118 |
| [66] |
BAUD S, DUBREUCQ B, MIQUEL M, ROCHAT C, LEPINIEC L. Storage reserve accumulation in Arabidopsis: Metabolic and developmental control of seed filling. The Arabidopsis Book, 2008,6:e0113.
doi: 10.1199/tab.0113 |
| [67] |
BUITINK J, LEPRINCE O. Intracellular glasses and seed survival in the dry state. Comptes Rendus Biologies, 2008,331:788-795.
doi: 10.1016/j.crvi.2008.08.002 |
| [68] |
WALTERS C. Orthodoxy, recalcitrance and in-between: Describing variation in seed storage characteristics using threshold responses to water loss. Planta, 2015,242:397-406.
doi: 10.1007/s00425-015-2312-6 |
| [69] | GONZÁLEZ-MORALES S I, CHÁVEZ-MONTES R A, HAYANO- KANASHIRO C, ALEJO-JACUINDE G, RICO-CAMBRON T Y, DE FOLTER S, HERRERA-ESTRELLA L. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United Stated America, 2016,113:E5232-E5241. |
| [70] |
JING Y, LANG S, WANG D, XUE H, WANG X F. Functional characterization of galactinol synthase and raffinose synthase in desiccation tolerance acquisition in developing Arabidopsis seeds. Journal of Plant Physiology, 2018,230:109-121.
doi: 10.1016/j.jplph.2018.10.011 |
| [71] |
HELL A F, KRETZSCHMAR F S, SIMÕES K, HEYER A G, BARBEDO C J, BRAGA M R, CENTENO D C. Metabolic changes on the acquisition of desiccation tolerance in seeds of the brazilian native tree Erythrina speciosa. Frontiers in Plant Science, 2019,10:1356.
doi: 10.3389/fpls.2019.01356 |
| [72] |
PUKACKA S, RATAJCZAK E, KALEMBA E. Non-reducing sugar levels in beech (Fagus sylvatica) seeds as related to withstanding desiccation and storage. Journal of Plant Physiology, 2009,166:1381-1390.
doi: 10.1016/j.jplph.2009.02.013 |
| [73] |
INGRAM J, CHANDLER J W, GALLAGHER L, SALAMINI F, BARTELS D. Analysis of cDNA clones encoding sucrose-phosphate synthase in relation to sugar interconversions associated with dehydration in the resurrection plant Craterostigma plantagineum Hochst. Plant Physiology, 1997,115:113-121.
doi: 10.1104/pp.115.1.113 |
| [74] |
PETERS S, MUNDREE S G, THOMSON J A, FARRANT J M, KELLER F. Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): Both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. Journal of Experimental Botany, 2007,58:1947-1956.
doi: 10.1093/jxb/erm056 |
| [75] |
YOSHIDA T, MOGAMI J, YAMAGUCHI-SHINOZAKI K. ABA- dependent and ABA-independent signaling in response to osmotic stress in plants. Current Opinion in Plant Biology, 2014,21:133-139.
doi: 10.1016/j.pbi.2014.07.009 |
| [76] |
LIU S, LÜ Z, LIU Y, LI L, ZHANG L. Network analysis of ABA-dependent and ABA- independent drought responsive genes in Arabidopsis thaliana. Genetics and Molecular Biology, 2018,41:624-637.
doi: 10.1590/1678-4685-gmb-2017-0229 |
| [77] |
FATIHI A, BOULARD C, BOUYER D, BAUD S, DUBREUCQ B, LEPINIEC L. Deciphering and modifying LAFL transcriptional regulatory network in seed for improving yield and quality of storage compounds. Plant Science, 2016,250:198-204.
doi: 10.1016/j.plantsci.2016.06.013 |
| [78] | CARBONERO P, IGLESIAS-FERNÁNDEZ R, VICENTE- CARBAJOSA J. The AFL subfamily of B3 transcription factors: Evolution and function in angiosperm seeds. Journal of Experimental Botany, 2017,68:871-880. |
| [79] |
YAMASAKI K, KIGAWA T, SEKI M, SHINOZAKI K, YOKOYAMA S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends in Plant Science, 2013,18:267-276.
doi: 10.1016/j.tplants.2012.09.001 |
| [80] | BRAYBROOK S A, STONE S L, PARK S, BUI A Q, LE B H, FISCHER R L, GOLDBERG R B, HARADA J J. Genes directly regulated by LEAFY COTYLEDON 2 provide insight into the control of embryo maturation and somatic embryogenesis. Proceedings of the National Academy of Sciences of the United Stated America, 2006,103:3468-3473. |
| [81] |
GRIMAULT A, GENDROT G, CHAIGNON S, GILARD F, TCHERKEZ G, THÈVENIN J, DUBREUCQ B, DEPÈGE-FARGEIK N, ROGOWSKY P M. Role of B3 domain transcription factors of the AFL family in maize kernel filling. Plant Science, 2015,236:116-125.
doi: 10.1016/j.plantsci.2015.03.021 |
| [82] |
TO A, VALON C, SAVINO G, GUILLEMINOT J, DRVIC M, GIRAUDAT J, PARCY F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. The Plant Cell, 2006,18:1642-1651.
doi: 10.1105/tpc.105.039925 |
| [83] | RIGHETTI K, VU J L, PELLETIER S, VU B L, GLAAB E, LALANNE D, PASHA A, PATEL R V, PROVART N J, VERDIER J, LEPRINCE O, BUITINKA J. Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. The Plant Cell, 2015,27:2692-2708. |
| [84] |
BIES-ETHÈVE N, GAUBIER-COMELLA P, DEBURES A, LASSERRE E, JOBET E, RAYNAL M, COOKE R, DELSENY M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Molecular Biology, 2008,67:107-124.
doi: 10.1007/s11103-008-9304-x |
| [85] |
CHEN K, LI G, BRESSAN R A, SONG C, ZHU J, ZHAO Y. Abscisic acid dynamics, signaling, and functions in plants. Journal of Integrative Plant Biology, 2020,62:25-54.
doi: 10.1111/jipb.v62.1 |
| [86] |
JO L, PELLETIER J M, HARADA J J. Central role of the LEAFY COTYLEDON 1 transcription factor in seed development. Journal of Integrative Plant Biology, 2019,61:564-580.
doi: 10.1111/jipb.v61.5 |
| [87] |
BAUD S, MENDOZA M S, TO A, HARSCOËT E, LEPINIEC L, DUBREUCQ B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON 2 towards fatty acid metabolism during seed maturation in Arabidopsis. The Plant Journal, 2007,50:825-838.
doi: 10.1111/j.1365-313X.2007.03092.x |
| [88] |
CHE N, YANG Y, LI Y, WANG L, HUANG P, GAO Y, AN C. Efficient LEC2 activation of OLEOSIN expression requires two neighboring RY elements on its promoter. Science in China Series C-Life Sciences, 2009,52:854-863.
doi: 10.1007/s11427-009-0119-z |
| [89] |
BRAYBROOK S A, HARADA J J. LECs go crazy in embryo development. Trends in Plant Science, 2008,13:624-630.
doi: 10.1016/j.tplants.2008.09.008 |
| [90] |
GRAEBER K, NAKABAYASHI K, MIATTON E, LEUBNER- METZGER G, SOPPE W J J. Molecular mechanisms of seed dormancy. Plant Cell and Environment, 2012,35:1769-1786.
doi: 10.1111/pce.2012.35.issue-10 |
| [91] |
WANG F, PERRY S E. Identification of direct targets of fusca3, a key regulator of Arabidopsis seed development. Plant Physiology, 2013,161:1251-1264.
doi: 10.1104/pp.112.212282 |
| [92] |
CHEN M, ZHANG B, LI C, KULAVEERASINGAM H, CHEW F T, YU H. TRANSPARENT TESTA GLABRA 1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiology, 2015,169:391-402.
doi: 10.1104/pp.15.00943 |
| [93] |
YAMAMOTO A, KAGAYA Y, USUI H, HOBO T, TAKEDA S, HATTORI T. Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell and Physiology, 2010,51:2031-2046.
doi: 10.1093/pcp/pcq162 |
| [94] | BERR A, SHEN W H. Molecular mechanisms in epigenetic regulation of plant growth and development//PUA E C, DAVEY M R, eds. Plant Developmental Biology Biotechnological Perspectives. Springer: Berlin/Heidelberg Press, 2010: 325-344. |
| [95] | CABEJ N R. Epigenetic Principles of Evolution. Elsevier Inc.: Amsterdam, The Netherlands, 2019: 733-781. |
| [96] |
PLITTA-MICHALAK B P, NASKRET-BARCISZEWSKA M Z, KOTLARSKI S, TOMASZEWSKI D, TYLKOWSKI T, BARCISZEWSKI J, CHMIELARZ P, MICHALAK M. Changes in genomic 5-methylcytosine level mirror the response of orthodox (Acer platanoides L.) and recalcitrant (Acer pseudoplatanus L.) seeds to severe desiccation. Tree Physiology, 2018,38:617-629.
doi: 10.1093/treephys/tpx134 |
| [97] |
LEBEDEVA M A, TVOROGOVA V E, TIKHODEYEV O N. Epigenetic mechanisms and their role in plant development. Russian Journal of Genetics, 2017,53:1057-1071.
doi: 10.1134/S1022795417090083 |
| [98] |
BOUYER D, KRAMDI A, KASSAM M, HEESE M, SCHNITTGER A, ROUDIER F, COLOT V. DNA methylation dynamics during early plant life. Genome Biology, 2017,18:1-12.
doi: 10.1186/s13059-016-1139-1 |
| [99] |
BARTELS A, HAN Q, NAIR P, STACEY L, GAYNIER H, MOSLEY M, HUANG Q, PEARSON J, HSIEH T F, AN Y Q, XIAO W. Dynamic DNA methylation in plant growth and development. International Journal of Molecular Science, 2018,19:2144.
doi: 10.3390/ijms19072144 |
| [100] |
KAWAKATSU T, NERY J R, CASTANON R, ECKER J R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biology, 2017,18:1-12.
doi: 10.1186/s13059-016-1139-1 |
| [101] | CHEN M, LIN J Y, HUR J, PELLETIER J M, BADEN R, PELLEGRINI M, HARADA J J, GOLDBERG R B. Seed genome hypomethylated regions are enriched in transcription factor genes. Proceedings of the National Academy of Sciences of the United States of America, 2018,115:E8315-E8322. |
| [102] |
AN Y Q C, GOETTEL W, HAN Q, BARTELS A, LIU Z, XIAO W. Dynamic changes of genome-wide DNA methylation during soybean seed development. Scientific Reports, 2017,7:1-14.
doi: 10.1038/s41598-016-0028-x |
| [103] |
MICHALAK M, BARCISZEWSKA M Z, BARCISZEWSKI J, PLITTA B P, CHMIELARZ P. Global changes in DNA methylation in seeds and seedlings of Pyrus communis after seed desiccation and storage. PLoS ONE, 2013,8:e70693.
doi: 10.1371/journal.pone.0070693 |
| [104] |
LI Y, KUMAR S, QIAN W. Active DNA demethylation: Mechanism and role in plant development. Plant Cell Report, 2018,37:77-85.
doi: 10.1007/s00299-017-2215-z |
| [105] |
ZHU J K. Active DNA demethylation mediated by DNA glycosylases. Annual Review of Genetics, 2009,43:143-166.
doi: 10.1146/genet.2009.43.issue-1 |
| [106] |
LIU R, LANG Z. The mechanism and function of active DNA demethylation in plants. Journal of Integrative Plant Biology, 2020,62:148-159.
doi: 10.1111/jipb.v62.1 |
| [107] |
LEPINIEC L, DEVIC M, ROSCOE T J, BOUYER D, ZHOU D X, BOULARD C, BAUD S, DUBREUCQ B. Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reproduction, 2018,31:291-307.
doi: 10.1007/s00497-018-0337-2 |
| [108] |
NONOGAKI H. Seed germination and dormancy: The classic story, new puzzles, and evolution. Journal of Integrative Plant Biology, 2019,61:541-563.
doi: 10.1111/jipb.v61.5 |
| [109] |
SALL K, DEKKERS B J W, NONOGAKI M, KATSURAGAWA Y, KOYARI R, HENDRIX D, WILLEMS L A J, BENTSINK L, NONOGAKI H. DELAY OF GERMINATION 1-LIKE 4 acts as an inducer of seed reserve accumulation. The Plant Journal, 2019,100:7-19.
doi: 10.1111/tpj.v100.1 |
| [110] |
SOPPE W J J, BENTSINK L. Seed dormancy back on track; its definition and regulation by DOG1. New Phytologist, 2020,228:816-819.
doi: 10.1111/nph.v228.3 |
| [111] |
GUTIERREZ L, WUYTSWINKEL O V, CASTELAIN M, BELLINI C. Combined networks regulating seed maturation. Trends in Plant Science, 2007,12:294-300.
doi: 10.1016/j.tplants.2007.06.003 |
| [112] |
NAKABAYASHI K, BARTSCH M, XIANG Y, MIATTON E, PELLENGAHR S, YANO R, SEO M, SOPPE W J J. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. The Plant Cell, 2012,24:2826-2838.
doi: 10.1105/tpc.112.100214 |
| [113] |
CUTLER S R, RODRIGUEZ P L, FINKELSTEIN R R, ABRAMS S R. Abscisic acid: Emergence of a core signaling network. Annual Review of Plant Biology, 2010,61:651-679.
doi: 10.1146/arplant.2010.61.issue-1 |
| [114] |
DEJONGHE W, OKAMOTO M, CUTLER S R. Small molecule probes of ABA biosynthesis and signaling. Plant Cell and Physiology, 2018,59:1490-1499.
doi: 10.1093/pcp/pcy126 |
| [115] |
XU P, CAI W. Function of Brassica napus BnABI3 in Arabidopsis gs1, an allele of AtABI3, in seed development and stress response. Frontiers in Plant Science, 2019,10:67.
doi: 10.3389/fpls.2019.00067 |
| [116] |
NISHIMURA N, TSUCHIYA W, MORESCO J J, HAYASHI Y, SATOH K, KAIWA N, IRISA T, KIOOSHITA T, SCHROEDER J I, YATES J R, HIRAYAMA T, YAMAZAKI T. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nature Communications, 2018,9:2132.
doi: 10.1038/s41467-018-04437-9 |
| [117] |
DEKKERS B J W, BENTSINK L. Regulation of seed dormancy by abscisic acid and delay of germination 1. Seed Science Research, 2015,25:82-98.
doi: 10.1017/S0960258514000415 |
| [118] |
MAIA J, DEKKERS B J W, DOLLE M J, LIGTERINK W, HILHORST H W M. Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytologist, 2014,203:81-93.
doi: 10.1111/nph.2014.203.issue-1 |
| [1] | 范子晗,罗雅尹,熊华烨,张育文,康福蓉,王昱桁,王洁,石孝均,张跃强. 酸性土壤硝化作用对柑橘铵毒害的效应[J]. 中国农业科学, 2022, 55(18): 3600-3612. |
| [2] | 尹飞,李振宇,SAMINA Shabbir,林庆胜. P450基因在氯虫苯甲酰胺不同抗性品系小菜蛾中的表达及功能分析[J]. 中国农业科学, 2022, 55(13): 2562-2571. |
| [3] | 戴思兰,洪艳. 基于花青素苷合成和呈色机理的观赏植物花色改良分子育种[J]. 中国农业科学, 2016, 49(3): 529-542. |
| [4] | 宋世佳,孙红春,张永江,刘连涛,白志英,李存东. 彩色棉抗氧化系统生理特征及纤维素累积对纤维品质的影响[J]. 中国农业科学, 2015, 48(19): 3811-3820. |
| [5] | 惠竹梅, 王智真, 胡勇, 邓敏敏, 张振文. 24-表油菜素内酯对低温胁迫下葡萄幼苗抗氧化系统 及渗透调节物质的影响[J]. 中国农业科学, 2013, 46(5): 1005-1013. |
| [6] | 张帆, 郁继华, 颉建明, 冯致, 张国斌, 李雯琳. 外源ALA和Spd对低温弱光下辣椒幼苗光合作用 及抗氧化系统的影响[J]. 中国农业科学, 2013, 46(11): 2298-2306. |
| [7] | 生利霞,冯立国,束怀瑞 . 低氧胁迫下钙对樱桃砧木根系抗氧化系统及线粒体功能的影响[J]. 中国农业科学, 2008, 41(11): 3913-3919 . |
| [8] | 康云艳,郭世荣,李 娟,段九菊. 24-表油菜素内酯对低氧胁迫下黄瓜幼苗根系抗氧化系统的影响[J]. 中国农业科学, 2008, 41(1): 153-161 . |
| [9] | 马纯艳,徐 昕,郝 林,曹 军. 小白菜幼苗对二氧化氮胁迫的应答及过氧化氢的调节[J]. 中国农业科学, 2007, 40(11): 2556-2562 . |
| [10] | 吴晓亮,辛萍萍,张志娥,陈晓玲,陶 澜,卢新雄. 水稻种子室温贮藏最适含水量及其热稳定蛋白的研究[J]. 中国农业科学, 2006, 39(11): 2214-2219 . |
| [11] | 陈贵林. 钙和钙调素拮抗剂对高温胁迫下茄子幼苗抗氧化系统的影响[J]. 中国农业科学, 2005, 38(01): 197-202 . |
| [12] | 汪炳良, 徐敏, 史庆华, 曹家树. 高温胁迫对早熟花椰菜叶片抗氧化系统和叶绿素及其荧光参数的影响[J]. 中国农业科学, 2004, 37(08): 1245-1245 . |
|
||