













中国农业科学 ›› 2025, Vol. 58 ›› Issue (11): 2062-2080.doi: 10.3864/j.issn.0578-1752.2025.11.002
曾跃辉1,2( ), 邹文广2,3(
), 邹文广2,3( ), 赵福铭1,2, 肖长春1,2, 黄建鸿1,2, 马彬林2,3, 杨旺兴2,3, 韦新宇1,2(
), 赵福铭1,2, 肖长春1,2, 黄建鸿1,2, 马彬林2,3, 杨旺兴2,3, 韦新宇1,2( ), 许旭明2,3(
), 许旭明2,3( )
)
收稿日期:2024-11-20
接受日期:2024-12-19
出版日期:2025-06-01
发布日期:2025-06-09
通信作者:
联系方式:
曾跃辉,E-mail:1_zengyuehui_1@163.com。邹文广,E-mail:52587656@qq.com。曾跃辉和邹文广为同等贡献作者。
基金资助:
ZENG YueHui1,2( ), ZOU WenGuang2,3(
), ZOU WenGuang2,3( ), ZHAO FuMing1,2, XIAO ChangChun1,2, HUANG JianHong1,2, MA BinLin2,3, YANG WangXing2,3, WEI XinYu1,2(
), ZHAO FuMing1,2, XIAO ChangChun1,2, HUANG JianHong1,2, MA BinLin2,3, YANG WangXing2,3, WEI XinYu1,2( ), XU XuMing2,3(
), XU XuMing2,3( )
)
Received:2024-11-20
Accepted:2024-12-19
Published:2025-06-01
Online:2025-06-09
摘要:
【目的】 水稻颖花开裂突变体sg2(split glume 2)来源于三明市农业科学研究院自主选育的恢复系GK1327高世代育种群体的自然突变,通过对其颖花开裂基因OsSG2(Oryza sativa Split Glume 2)进行遗传学分析、图位克隆和功能验证,为进一步开展OsSG2在水稻花器官发育中的功能研究奠定基础和提供重要种质资源。【方法】 通过对突变体sg2颖花进行形态特征解剖观察,分析其花器官发育突变特点。通过对野生型WT(GK1327)与突变体sg2表型和农艺性状进行调查,以及花粉在I2-KI溶液中的染色观察,分析该突变表型对主要产量性状和花粉育性的影响。利用突变体sg2分别与野生型WT和9311杂交,构建多个F2分离群体,对OsSG2进行遗传学分析和物理定位。采用集群分离分析法(bulked segregant analysis,BSA)和二代测序(next-generation sequencing,NGS)相结合的BSA-seq技术,以及半定量PCR和实时荧光定量PCR对候选基因进行筛选和表达模式分析。结合PCR扩增和测序技术对候选基因进行克隆和序列分析。通过CRISPR-Cas9基因编辑技术和转基因互补试验对OsSG2候选基因进行功能验证。【结果】 该突变体在生殖生长时期花器官发育出现明显异常,其颖花内外稃片瘪弱,表现出不同程度的扭曲变形,且开裂不抱合表型,穗部颖花发育不正常的比例达到100%。相比野生型,突变体sg2结实率和千粒重显著降低,种子萌发率显著下降,花粉育性未出现明显异常。遗传分析表明,sg2突变表型由单个细胞核隐性基因控制。通过图位克隆技术,将OsSG2精细定位在水稻第3染色体短臂端,位于2个InDel(insertion/deletion)标记InDel-12083和InDel-17610之间,物理距离为919.85 kb,该区间共注释75个开放阅读框(open read frames,ORFs)。利用BSA-seq技术,鉴定到一个潜在的OsSG2候选基因LOC_Os03g11614,该基因编码一个定位于细胞核的OsMADS1转录因子。表达和序列分析表明,相比野生型,LOC_Os03g11614在突变体sg2抽穗期幼穗中的表达水平极显著下调,且该基因编码序列(coding sequence,CDS)在突变体中发生单个核苷酸多态性(single nucleotide polymorphism,SNP)突变,即LOC_Os03g11614第1外显子第3个碱基由G突变为A,导致其起始密码子由ATG突变为ATA,进而影响该基因的转录表达和相应蛋白OsMADS1的结构和功能。CRISPR-Cas9基因编辑技术和功能互补试验进一步证实候选基因LOC_Os03g11614即为OsSG2。【结论】 鉴定并克隆获得的水稻颖花开裂基因OsSG2为一个新的OsMADS1等位突变基因。
曾跃辉, 邹文广, 赵福铭, 肖长春, 黄建鸿, 马彬林, 杨旺兴, 韦新宇, 许旭明. 一个新的水稻颖花开裂基因OsSG2的图位克隆与功能验证[J]. 中国农业科学, 2025, 58(11): 2062-2080.
ZENG YueHui, ZOU WenGuang, ZHAO FuMing, XIAO ChangChun, HUANG JianHong, MA BinLin, YANG WangXing, WEI XinYu, XU XuMing. Map-Based Cloning and Functional Verification of A Novel Split Glume Gene OsSG2 in Rice (Oryza sativa L.)[J]. Scientia Agricultura Sinica, 2025, 58(11): 2062-2080.
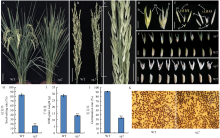
图1
野生型与突变体sg2的表型和农艺性状 A:野生型和突变体sg2灌浆期单株表型,Bar=10 cm;B—C:野生型和突变体sg2灌浆期穗子表型,Bar=5 cm,其中,图C为图B突变体sg2穗子局部放大视野,Bar=2 cm;D:突变体sg2抽穗期颖花表型,Bar=1 cm;E:突变体sg2颖花结构解剖,Bar=1 cm;F—G:野生型和突变体sg2灌浆期(F,Bar=1 cm)和成熟期(G,Bar=1 cm)籽粒表型;H—J:野生型和突变体sg2结实率(H)、千粒重(I)以及种子发芽率(J);K:野生型和突变体sg2花粉育性I2-KI染色鉴定,Bar=1 mm。le:外稃;pa:内稃;st:雄蕊;pi:雌蕊;sl:护颖;WT:GK1327。**:P<0.01。下同"

表2
本研究所使用的引物序列"
| 引物名称 Primer name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) | 用途 Usage |
|---|---|---|---|
| Rm14462 | TCTGTATTCAAGGAGGGCTAGATGC | GCTGGAGAGATGCATTGACTGG | OsSG2初步定位 Preliminary mapping for OsSG2 |
| Rm3392 | AGCAACCAACCCAGTAGTTAGCC | GCTCATTTGCATGCTGTGTTAGC | |
| Rm14722 | ACACACCACATTGTTGGCATTGG | TGGTGGGAGGAAGATTTCAGTGG | |
| Rm14728 | CTCAGCGGACTAGAATGCAAGC | AAACGGATACATCTTCGCAGAACG | |
| Rm7 | TTCGCCATGAAGTCTCTCG | CCTCCCATCATTTCGTTGTT | |
| InDel-2923 | GCCTAAATTTGCAGTGATGTG | TACCAGTGCCCAATAATCGT | OsSG2精细定位 Fine mapping for OsSG2 |
| InDel-12083 | GCAATCAGGTCGCGTATCAG | GGTTGGGTGTTCGACGTATC | |
| InDel-17610 | ACGAATGTAACTTTGGCATGTCA | ATGCTCTGAGGTGATTACTTTT | |
| InDel-31068 | AAGCCCCGTTTGACTTAGGT | AAGCCCCGTTTGACTTAGGT | |
| InDel-31355 | TTAGGCCCAGTTTTGCTCTC | CTCCCTCCGTCCCGAAAA | |
| OsMADS1-Q | TTGGAGAGGTACCGCAGC | TCTGGTTCTCAAGCTGCTCC | OsMADS1的qRT-PCR检测 qRT-PCR detection for OsMADS1 |
| OsMADS1-Seq01 | CGACGTGACAGCAGTTTGAT | AGCACTAAAAGCACGCGAAA | OsMADS1的测序 Sequencing for OsMADS1 |
| OsMADS1-Seq02 | TGCGCAAATCAAAACTGTACAG | ATTGCGGCCCATCTAAACTG | |
| OsMADS1-Seq03 | TAGGGCGCATTGTGACTTTG | GCCTGTGTTAAAAGCCATGC | |
| OsMADS1-Seq04 | AGCAAGCACTTCACACCATG | AGAACAGTGCTAGAACCGCT | |
| OsMADS1-Seq05 | AGCCATCGATCACCCTGAAA | GTCTGCTGCTTCATTGCTCA | |
| OsMADS1-Seq06 | CCATCAGGGTCTTCTCCACC | TGAGATAACAGCTGCCGTCT | |
| OsActin-F/R | ACCTTCAACACCCCTGCTAT | CACCATCACCAGAGTCCAAC | OsActin的qRT-PCR检测 qRT-PCR detection for OsActin (as internal standard) |
| FMHPT-F/R | ATTTGTGTACGCCCGACAGT | GTGCTTGACATTGGGGAGTT | PCR鉴定HPT PCR detection for HPT |
| FMCas9-F/R | AGAACCTCTCCGATGCTATCC | AGCAAGAGGACCAACGTAG | PCR鉴定Cas9 PCR detection for Cas9 |
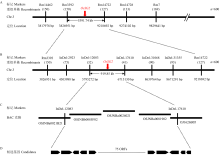
图2
OsSG2的精细定位 A:OsSG2初步定位在第3染色体且位于SSR分子标记Rm3392和Rm14722之间;B:利用9311与sg2杂交(9311×sg2、sg2×9311)获得的共600个F2突变单株将OsSG2精细定位在InDel标记InDel-12083和InDel-17610之间,区间物理距离为919.85 kb;C:包含OsSG2位点的BAC克隆重叠群;D:参考粳稻品种日本晴基因组信息,定位区间共注释有75个ORFs作为OsSG2的候选基因。与OsSG2连锁的各分子标记下方括号中的数字表示从600个F2突变单株中鉴定到的重组交换单株。染色体下方数字表示相应连锁分子标记在水稻第3染色体上的实际物理位置"

表4
测序深度和覆盖度"
| 样本 Samples | 有效序列数 Total reads | 比对序列数 Mapped reads | 比对率 Mapped rate (%) | 平均覆盖深度 Average coverage (×) | 1×覆盖度 1×coverage (%) | 10×覆盖度 10×coverage (%) |
|---|---|---|---|---|---|---|
| 9311 | 176181050 | 136126271 | 98.23 | 52.7028 | 93.72 | 88.85 |
| sg2 | 182366852 | 140044150 | 98.16 | 54.2271 | 93.16 | 88.38 |
| WT-Pool | 192599862 | 145760573 | 98.13 | 56.2530 | 94.71 | 90.18 |
| MT-Pool | 201645008 | 152345579 | 98.11 | 58.7479 | 94.74 | 90.45 |
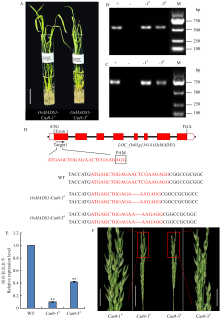
图6
OsSG2候选基因LOC_Os03g11614的CRISPR-Cas9功能验证 A:利用CRISPR-Cas9基因编辑系统获得的2个T0代阳性转基因株系OsMADS1-Cas9-1#和OsMADS1-Cas9-3#,Bar=3 cm;B—C:功能标记FMCas9-F/R(B)和FMHPT-F/R(C)对T0代阳性转基因植株PCR检测,+和-分别表示阳性对照和阴性对照,M代表2000 bp DNA Marker;D:OsMADS1-Cas9-1#和OsMADS1-Cas9-3#中LOC_Os03g11614靶点1突变类型分析,红色字体表示靶点序列,黑色方框中的核苷酸表示PAM序列,-表示碱基缺失;E:qRT-PCR分析LOC_Os03g11614分别在野生型、OsMADS1-Cas9-1#和OsMADS1-Cas9-3#抽穗期幼穗中的表达水平;F:OsMADS1-Cas9-1#和OsMADS1-Cas9-3#抽穗期幼穗表型,标尺表示2 cm(左)、6 cm(中)和2 cm(右)。WT:GK1327"

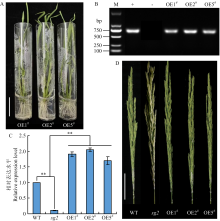
图7
OsSG2候选基因LOC_Os03g11614的功能互补验证 A:利用互补试验获得的3个T0代阳性转基因株系OsMADS1-OE1#、OsMADS1-OE2#和OsMADS1-OE5#,Bar=3 cm;B:功能标记FMHPT-F/R对T0代阳性过表达转基因植株PCR检测,+和-分别表示阳性对照和阴性对照,M代表2000 bp DNA Marker;C:qRT-PCR分析LOC_Os03g11614分别在野生型、突变体sg2、OsMADS1-OE1#、OsMADS1-OE2#和OsMADS1-OE5#抽穗期幼穗中的表达水平;D:OsMADS1-OE1#、OsMADS1-OE2#和OsMADS1-OE5#抽穗期幼穗表型,Bar=6 cm。WT:GK1327"

| [1] |
|
| [2] |
陈晓东, 李飞, 李丹阳, 刘灵芝, 乔守晨, 孙红正, 李俊周, 张静, 彭廷, 谭金芳, 杜彦修, 赵全志. 水稻内颖畸形或缺失基因OsAPP1的图位克隆及表达分析. 植物遗传资源学报, 2021, 22(2): 416-426.
doi: 10.13430/j.cnki.jpgr.20200901001 |
|
|
|
| [3] |
|
| [4] |
曾生元, 郭旻, 李荣德, 盛生兰, 龚红兵, 严长杰. 水稻新裂颖突变体sg1的遗传分析与基因定位. 中国水稻科学, 2015, 29(1): 22-27.
doi: 10.3969/j.issn.1001-7216.2015.01.003 |
|
|
|
| [5] |
潘孝武, 刘文强, 黎用朝, 熊海波, 盛新年, 段永红, 余亚莹, 赵文锦, 魏秀彩, 李小湘. 水稻裂颖突变体sh1的鉴定及基因定位. 中国水稻科学, 2019, 33(4): 323-330.
doi: 10.16819/j.1001-7216.2019.9027 |
|
|
|
| [6] |
姜树坤, 张喜娟, 王嘉宇, 张凤鸣. 水稻幼穗-颖花发育的研究进展. 植物遗传资源学报, 2012, 13(6): 1018-1022, 1030.
doi: 10.13430/j.cnki.jpgr.2012.06.016 |
|
|
|
| [7] |
|
| [8] | |
|
|
|
| [9] |
管恩相, 李勇, 刘陵武, 姚仁祥, 郭君, 唐海燕. 三系不育系种子裂颖形成的原因及防治措施研究. 中国种业, 2010(1): 42-43.
|
|
|
|
| [10] |
朱伟, 童继平, 吴跃进. 水稻品种抗裂颖资源的筛选与初步研究. 植物遗传资源学报, 2004, 5(1): 52-55.
|
|
|
|
| [11] |
王忠, 顾蕴洁, 高煜珠.水稻开颖机理的探讨: V. 不育系与可育系浆片和花丝结构的比较. 作物学报, 1994, 20(1): 13-17, 129-131.
|
|
|
|
| [12] |
doi: 10.1016/0092-8674(94)90291-7 pmid: 7913881 |
| [13] |
|
| [14] |
doi: 10.1016/S0092-8674(00)80618-2 pmid: 10778850 |
| [15] |
doi: 10.1016/s1369-5266(00)00139-4 pmid: 11163172 |
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
doi: 10.1093/aob/mcm143 pmid: 17670752 |
| [20] |
doi: 10.1093/jxb/err272 pmid: 21914655 |
| [21] |
|
| [22] |
|
| [23] |
doi: 10.1093/jxb/erz198 pmid: 31034557 |
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
doi: 10.1093/jxb/eraa588 pmid: 33337484 |
| [31] |
doi: 10.1093/jxb/eraa460 pmid: 33035313 |
| [32] |
|
| [33] |
doi: 10.1111/j.1365-313X.2005.02504.x pmid: 16146529 |
| [34] |
|
| [35] |
|
| [36] |
doi: 10.1016/S1672-6308(13)60112-2 |
| [37] |
doi: 10.1111/j.1365-313X.2009.04101.x pmid: 20003164 |
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
韦新宇, 曾跃辉, 杨旺兴, 肖长春, 候新坡, 黄建鸿, 邹文广, 许旭明. 利用CRISPR-Cas9技术编辑Badh2基因创制优质香型籼稻三系不育系. 作物学报, 2023, 49(8): 2144-2159.
doi: 10.3724/SP.J.1006.2023.22043 |
|
|
|
| [42] |
|
| [43] |
陈立凯, 黄明, 刘永柱, 王慧, 陈志强, 郭涛. 水稻开颖半不育突变体的观察、遗传分析和基因定位. 中国农业科学, 2016, 49(1): 1-13. doi: 10.3864/j.issn.0578-1752.2016.01.001.
|
|
|
|
| [44] |
孙廉平, 张迎信, 张沛沛, 杨正福, 占小登, 沈希宏, 张振华, 胡霞, 轩丹丹, 吴玮勋, 曹立勇, 程式华. 一个由可变剪接造成的水稻开颖不育突变体ohms1的鉴定及基因定位. 中国水稻科学, 2015, 29(5): 457-466.
doi: 10.3969/j.issn.1001G7216.2015.05.002 |
|
|
|
| [45] |
|
| [46] |
|
| [47] |
|
| [1] | 王爱冬, 李瑞杰, 冯向前, 洪卫源, 李子秋, 张晓果, 王丹英, 陈松. 基于多角度成像与机器学习的水稻叶面积精确估算[J]. 中国农业科学, 2025, 58(9): 1719-1734. |
| [2] | 韦萍, 潘炬忠, 朱德平, 邵胜雪, 陈珊珊, 韦雅倩, 高维维. OsDREB1J调控水稻籽粒大小的功能研究[J]. 中国农业科学, 2025, 58(8): 1463-1478. |
| [3] | 汪炜檬, 魏云晓, 唐云霓, 刘苗苗, 陈全家, 邓晓娟, 张锐. 棉花发根农杆菌转化体系的建立及生根优化[J]. 中国农业科学, 2025, 58(8): 1479-1493. |
| [4] | 刘劲松, 伍龙梅, 包晓哲, 刘志霞, 张彬, 杨陶陶. 短期减施氮肥对华南地区早晚兼用型水稻产量和稻米品质的影响[J]. 中国农业科学, 2025, 58(8): 1508-1520. |
| [5] | 王彬, 吴朋浩, 鲁剑巍, 任涛, 丛日环, 陆志峰, 李小坤. 长江中游地区水稻-油菜轮作体系需水特征[J]. 中国农业科学, 2025, 58(7): 1355-1365. |
| [6] | 罗刚, 程依依, 杨雯, 肖怡梦, 杨铖熹. CRISPR-Cas12a基因编辑技术及其在农业生产中的应用[J]. 中国农业科学, 2025, 58(7): 1434-1450. |
| [7] | 熊嘉妮, 李宗玥, 胡衡亮, 顾天宇, 高艳, 彭佳师. OsLCT1启动子驱动OsNRAMP5表达对镉向水稻种子迁移的影响[J]. 中国农业科学, 2025, 58(7): 1259-1268. |
| [8] | 晋艺丹, 何旎清, 程朝平, 林少俊, 黄凤凰, 白康呈, 张涛, 汪文潇, 余敏祥, 杨德卫. 稻瘟病抗病蛋白Pigm-1互作蛋白的筛选与鉴定[J]. 中国农业科学, 2025, 58(6): 1043-1051. |
| [9] | 靳亚茹, 陈斌, 王歆凯, 周田田, 李笑, 邓晶晶, 杨郁文, 郭冬姝, 张保龙. 利用基因编辑产生长片段缺失创制低谷蛋白水稻种质[J]. 中国农业科学, 2025, 58(6): 1052-1064. |
| [10] | 徐以恒. “基因编辑农作物”专利规制的困境及出路[J]. 中国农业科学, 2025, 58(5): 831-839. |
| [11] | 徐元元, 贾东升, 宾羽, 魏太云. 黑尾叶蝉PGRP6负调控共生菌抑制水稻矮缩病毒的经卵传播[J]. 中国农业科学, 2025, 58(5): 907-917. |
| [12] | 陈鸽, 谷雨, 文炯, 傅岳峰, 何兮, 李薇, 周峻宇, 刘琼峰, 吴海勇. 冬闲杂草还田对水稻光合物质生产和产量的影响[J]. 中国农业科学, 2025, 58(4): 647-659. |
| [13] | 王少骅, 沈年桥, 储天然, 吴永汉, 李康宁, 石延霞, 谢学文, 李磊, 范腾飞, 李宝聚, 柴阿丽. 浙江苍南番茄-水稻轮作对连作土壤理化性质和微生物群落的影响[J]. 中国农业科学, 2025, 58(4): 692-703. |
| [14] | 李璐, 谢庄, 谢可盈, 张瀚, 赵卓文, 向奥妮, 李巧龙, 凌英华, 何光华, 赵芳明. 水稻CSSL-Z492单、双片段代换系构建及粒型QTL的遗传解析[J]. 中国农业科学, 2025, 58(3): 401-415. |
| [15] | 赵守帅, 都梦翔, 郭思宇, 张烁, 赵宏伟, 赵长江. 钙调节活性氧和内生细菌增强水稻对纹枯病抗性[J]. 中国农业科学, 2025, 58(11): 2145-2161. |
|
||