













中国农业科学 ›› 2023, Vol. 56 ›› Issue (14): 2798-2811.doi: 10.3864/j.issn.0578-1752.2023.14.014
童雄( ), 罗威(
), 罗威( ), 闵力, 张志飞, 马新燕, 罗成龙, 陈卫东, 徐斌(
), 闵力, 张志飞, 马新燕, 罗成龙, 陈卫东, 徐斌( ), 李大刚(
), 李大刚( )
)
收稿日期:2023-01-04
接受日期:2023-04-23
出版日期:2023-07-16
发布日期:2023-07-21
通信作者:
联系方式:
童雄,E-mail:tongxiong@gdaas.cn。罗威,E-mail:luowei@gdaas.cn。童雄和罗威为同等贡献作者。
基金资助:
TONG Xiong( ), LUO Wei(
), LUO Wei( ), MIN Li, ZHANG ZhiFei, MA XinYan, LUO ChengLong, CHEN WeiDong, XU Bin(
), MIN Li, ZHANG ZhiFei, MA XinYan, LUO ChengLong, CHEN WeiDong, XU Bin( ), LI DaGang(
), LI DaGang( )
)
Received:2023-01-04
Accepted:2023-04-23
Published:2023-07-16
Online:2023-07-21
摘要:
【目的】研究陆丰黄牛和雷琼牛与世界不同地域家牛中的系统发育关系,解析不同家牛群体的遗传多样性,为地方家牛资源的鉴定与保护奠定理论基础。【方法】采集12头陆丰黄牛和17头雷琼牛的组织样品进行全基因组重测序,整合世界范围内分布的24个品种92个体的NCBI公共基因组数据,共计25个品种121个个体的信息开展群体遗传学研究。选用Bos taurus ARS-UCD1.2作为参考基因组,经基因组序列比对与质量控制获取高质量Reads,应用GATK软件检测基因组SNPs。进一步基于群体SNPs,利用系统进化树构建、PCA聚类和Admixture评估进行群体结构分析,通过核苷酸多样性(Pi)、杂合度(Hp)和连锁不平衡(LD)水平研究群体的遗传多样性。【结果】29头广东地方牛经基因组测序获取6 905 944 306个Clean Reads,每个样本平均基因组覆盖率为97.99%,平均测序深度分别为12.78×。整合NCBI公共基因组数据,经质控后共检测出14 664 391个群体SNPs。整合系统进化树、PCA、Admixture的结果发现普通牛与瘤牛分化,瘤牛群体存在中国瘤牛与印度瘤牛分化,普通牛群体中东北亚普通牛(韩牛、延边牛)和西藏牛与欧洲普通牛分化,温岭高峰牛和舟山牛从中国瘤牛群体中分化。陆丰黄牛和雷琼牛均属于纯正的中国瘤牛,但陆丰黄牛与皖南牛之间、雷琼牛与吉安牛之间呈现最近的亲缘关系,说明陆丰黄牛与地域临近的雷琼牛属于两个独立的品种。部分陆丰黄牛与雷琼牛均存在欧洲普通牛和东北亚普通牛的血统混杂,且混杂比例较高,说明这两个品种牛急需加强群体内的提纯复壮。相较于欧洲普通牛和韩牛,中国家牛群体的LD衰减速率更快,核苷酸多样性Pi和杂合度Hp更高,说明其遗传多样性更丰富。相较于其他中国家牛,陆丰黄牛和雷琼牛的LD水平更低,杂合度Hp更高,且核苷酸多样性Pi和杂合度Hp的密度分布更为集中,说明两者受人工选择强度较低,且维持较高的群体遗传多样性。【结论】通过全基因组SNPs标记,系统解析了陆丰黄牛与雷琼牛的群体遗传结构和多样性特征,为这两个品种独立分类及其保护利用提供数据支撑。
童雄, 罗威, 闵力, 张志飞, 马新燕, 罗成龙, 陈卫东, 徐斌, 李大刚. 基于全基因组SNPs分析陆丰黄牛和雷琼牛的群体结构与遗传多样性特征[J]. 中国农业科学, 2023, 56(14): 2798-2811.
TONG Xiong, LUO Wei, MIN Li, ZHANG ZhiFei, MA XinYan, LUO ChengLong, CHEN WeiDong, XU Bin, LI DaGang. Population Structure and Genetic Diversity of Lufeng Cattle and Leiqiong Cattle Based on Genome-Wide SNPs[J]. Scientia Agricultura Sinica, 2023, 56(14): 2798-2811.
表1
本研究中25个品种121个个体的样本信息"
| 品种 Breed | 品种代号 Breed code | 样本量 Sample size | 原产地 Origin region | 数据来源 Reference | 地理区域 Geographic region |
|---|---|---|---|---|---|
| 安格斯牛 Angus cattle | Angus | 5 | 英国苏格兰阿伯丁郡 Aberdeenshire, Scotland, UK | STOTHARD et al, 2015[ | 西欧 West Europe |
| 海福特牛 Hereford cattle | Hereford | 6 | 英国赫里福德郡 Hereford, UK | STOTHARD et al, 2015[ | 西欧 West Europe |
| 利木赞牛 Limousin cattle | Limousin | 1 | 法国利穆赞大区 Limousin, France | STOTHARD et al, 2015[ | 中南欧 Central-South Europe |
| 西门塔尔牛 Simmental cattle | Simmental | 6 | 瑞士日内瓦 Geneve, Switzerland | STOTHARD et al, 2015[ | 中南欧 Central-South Europe |
| 西藏牛 Tibetan cattle | Tibetan | 6 | 中国西藏自治区昌都市 Changdu, Xizang, China | CHEN et al, 2018[ | 西藏 Tibet |
| 韩牛 Hanwoo cattle | Hanwoo | 6 | 韩国江原道春川市 Chuncheon, Kangwon-do, Korea | SHIN et al, 2014[ | 东北亚 Northeast Asia |
| 延边牛 Yanbian cattle | Yanbian | 1 | 中国吉林省延边朝鲜族自治州 Yanbian, Jilin, China | CHEN et al, 2018[ | 东北亚 Northeast Asia |
| 鲁西牛 Luxi cattle | Luxi | 5 | 中国山东省济宁市 Jining, Shandong, China | CHEN et al, 2018[ | 中国中北部 North-Central China |
| 渤海黑牛 Bohai Black cattle | Bohai | 4 | 中国山东省滨州市 Binzhou, Shandong, China | CHEN et al, 2018[ | 中国中北部 North-Central China |
| 南阳牛 Nanyang cattle | Nanyang | 4 | 中国河南省南阳市 Nanyang, Henan, China | CHEN et al, 2018[ | 中国中北部 North-Central China |
| 锦江牛 Jinjiang cattle | Jinjiang | 3 | 中国江西省高安市 Gaoan, Jiangxi, China | CHEN et al, 2018[ | 中国南部 South China |
| 皖南牛 Wannan cattle | Wannan | 5 | 中国安徽省宣城市 Xuancheng, Anhui, China | CHEN et al, 2018[ | 中国南部 South China |
| 广丰牛 Guangfeng cattle | Guangfeng | 4 | 中国江西省上饶市 Shangrao, Jiangxi, China | CHEN et al, 2018[ | 中国南部 South China |
| 吉安牛 Ji’an cattle | Jian | 4 | 中国江西省吉安市 Ji’an, Jiangxi, China | CHEN et al, 2018[ | 中国南部 South China |
| 雷琼牛 Leiqiong cattle | Leiqiong | 3 | 中国广东省湛江市 Zhanjiang, Guangdong, China | CHEN et al, 2018[ | 中国南部 South China |
| Leiqiong | 17 | 中国广东省湛江市 Zhanjiang, Guangdong, China | 本研究 Present study | 中国南部 South China | |
| 陆丰黄牛 Lufeng cattle | Lufeng | 12 | 中国广东省汕尾市 Shanwei, Guangdong, China | 本研究 Present study | 中国南部 South China |
| 文山牛 Wenshan cattle | Wenshan | 6 | 中国云南省文山壮族苗族自治州 Wenshan, Yunnan, China | CHEN et al, 2018[ | 中国南部 South China |
| 舟山牛 Zhoushan cattle | Zhoushan | 6 | 中国浙江省舟山市 Zhoushan, Zhejiang, China | JIANG et al, 2021[ | 中国南部 South China |
| 温岭高峰牛 Wenling Hump cattle | Wenling | 6 | 中国浙江省温岭市 Wenling, Zhejiang, China | JIANG et al, 2021[ | 中国南部 South China |
| 哈里亚娜牛 Hariana cattle | Hariana | 1 | 印度哈里亚纳邦古尔加翁县 Gurgaon, Haryana, India | CHEN et al, 2018[ | 印巴地区 India-Paksitan |
| 吉尔牛 Gir cattle | Gir | 2 | 印度古吉拉特邦艾哈迈达巴德 Amblash, Gujarat, India | BICKHART et al, 2016[ | 印巴地区 India-Paksitan |
| 沙希华牛 Sahiwal cattle | Sahiwal | 1 | 巴基斯坦旁遮普省萨希瓦尔 Sahiwal, Punjab, Pakistan | CHEN et al, 2018[ | 印巴地区 India-Paksitan |
| 内洛尔牛 Nelore cattle | Nelore | 1 | 印度安得拉邦内洛尔县 Nelore, Andhra, India | BICKHART et al, 2016[ | 印巴地区 India-Paksitan |
| 塔帕卡牛 Tharparkar cattle | Tharparkar | 1 | 巴基斯坦信德省塔帕卡 Tharparkar, Sindh, Pakistan | CHEN et al, 2018[ | 印巴地区 India-Paksitan |
| 婆罗门牛 Brahman cattle | Brahman | 5 | 印度西孟加拉邦婆罗门 Brahman, West Bengal, India | BICKHART et al, 2016[ | 印巴地区 India-Paksitan |
| 总计 Total | 25品种 25 breeds | 121个体 121 individuals | |||
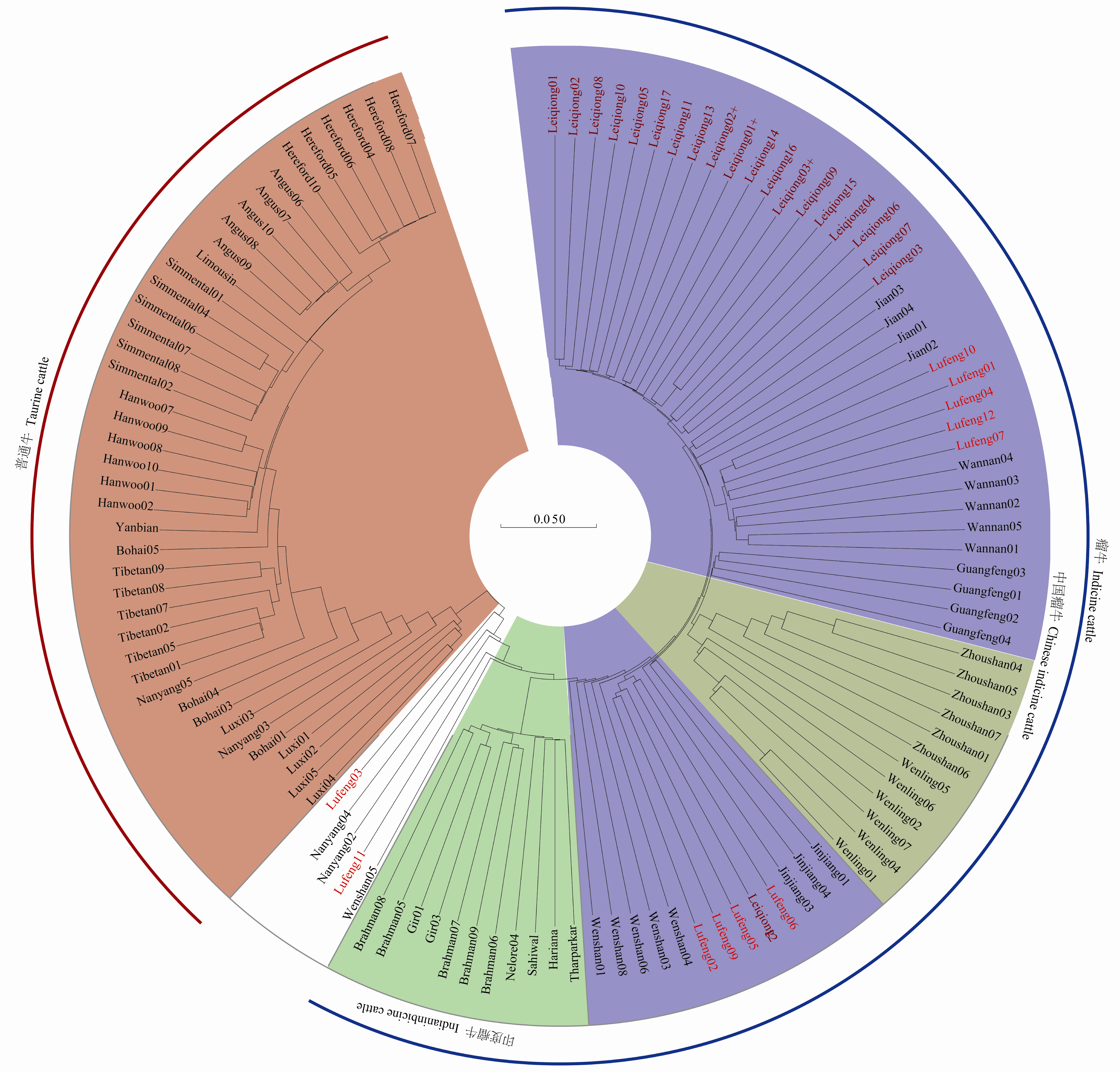
图3
基于IBS距离构建不同地域家牛的邻近进化树 Angus:安格斯牛;Hereford:海福特牛;Limousin:利木赞牛;Simmental:西门塔尔牛;Tibetan:西藏牛;Hanwoo:韩牛;Yanbian:延边牛;Luxi:鲁西牛;Bohai:渤海黑牛;Nanyang:南阳牛;Jinjiang:锦江牛;Wannan:皖南牛;Guangfeng:广丰牛;Jian:吉安牛;Leiqiong:雷琼牛;Lufeng:陆丰黄牛;Wenshan:文山牛;Zhoushan:舟山牛;Wenling:温岭高峰牛;Hariana:哈里亚娜牛;Gir:吉尔牛;Sahiwal:沙希华牛;Nelore:内洛尔牛;Tharparkar:塔帕卡牛;Brahman:婆罗门牛。品种代号后的数字代表品种内个体的编号。下同"
| [1] |
|
| [2] |
|
| [3] |
国家畜禽遗传资源委员会组. 中国畜禽遗传资源志-牛志. 北京: 中国农业出版社, 2011.
|
|
China National Commission of Animal Genetic Resources. Animal genetic resources in China. Beijing: China Agriculture Press, 2011. (in Chinese)
|
|
| [4] |
广东省农业厅,广东省畜牧兽医局, 广东省畜禽遗传资源委员会. 广东省地方畜禽遗传资源志. 广州: 广东科技出版社, 2018.
|
|
Department of Agriculture of Guangdong Province. Records of local livestock and poultry genetic resources in Guangdong Province. Guangzhou: Guangdong Science & Technology Press, 2018. (in Chinese)
|
|
| [5] |
doi: 10.1111/age.12749 pmid: 30421479 |
| [6] |
doi: 10.1038/s41598-016-0028-x |
| [7] |
doi: 10.1038/s41467-017-02088-w |
| [8] |
doi: 10.1093/molbev/msx322 pmid: 29294071 |
| [9] |
俞英, 文际坤, 朱芳贤, 赵开典, 宿兵, 王文, 张亚平, 刘爱华, 季维智, 施立明. 云南文山黄牛和迪庆黄牛遗传多样性的蛋白电泳研究. 动物学研究, 1997, 18(3): 333-339.
|
|
|
|
| [10] |
王朝锋, 雷初朝, 陈宏, 蔡欣, 党瑞华, 欧江涛. 雷琼牛mtDNA D—loop遗传多态性研究. 黄牛杂志, 2005(5): 14-15.
|
|
|
|
| [11] |
pmid: 10467698 |
| [12] |
耿荣庆, 常洪, 冀德君, 王兰萍, 常国斌, 李世平, 马国龙, 陈宏宇, 常春芳, 李永红. 雷琼牛母系起源的遗传学证据. 畜牧兽医学报, 2008, 39(7)849-852.
|
|
|
|
| [13] |
doi: 10.3389/fgene.2020.533052 |
| [14] |
doi: 10.1371/journal.pone.0271718 |
| [15] |
doi: 10.1038/s41598-020-79139-8 |
| [16] |
doi: 10.1016/j.ygeno.2021.03.023 pmid: 33771637 |
| [17] |
doi: 10.1038/s41588-020-0694-2 pmid: 32989325 |
| [18] |
宋娜娜, 钟金城, 柴志欣, 汪琦, 何世明, 吴锦波, 蹇尚林, 冉强, 蒙欣. 三江黄牛全基因组数据分析. 中国农业科学, 2017, 50(1): 183-194.
doi: 10.3864/j.issn.0578-1752.2017.01.016 |
|
doi: 10.3864/j.issn.0578-1752.2017.01.016 |
|
| [19] |
doi: 10.1093/gigascience/giaa021 |
| [20] |
doi: 10.1093/bioinformatics/btp324 pmid: 19451168 |
| [21] |
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [22] |
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [23] |
doi: 10.1093/bioinformatics/btr330 pmid: 21653522 |
| [24] |
doi: 10.1086/519795 |
| [25] |
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [26] |
doi: 10.1371/journal.pgen.0020190 pmid: 17194218 |
| [27] |
doi: 10.1101/gr.094052.109 pmid: 19648217 |
| [28] |
doi: 10.1093/bioinformatics/bty875 pmid: 30321304 |
| [29] |
|
| [30] |
doi: 10.1186/1471-2164-15-240 |
| [31] |
doi: 10.1093/dnares/dsw013 pmid: 27085184 |
| [32] |
doi: 10.1371/journal.pone.0005753 |
| [33] |
doi: 10.1371/journal.pgen.1004254 |
| [34] |
doi: 10.1016/j.jas.2013.09.003 |
| [35] |
doi: 10.1017/S101423390000047X |
| [36] |
魏霞, 刘正刚. 明清安徽与广东的贸易往来. 安徽史学, 2001(4): 18-21.
|
|
|
|
| [37] |
王海. 明清粤赣通道与两省毗邻山地互动发展研究[D]. 广州: 暨南大学, 2008.
|
|
|
|
| [38] |
doi: 10.1186/s12864-020-07340-0 pmid: 33421990 |
| [39] |
doi: 10.1186/s12864-022-08645-y pmid: 35729510 |
| [40] |
doi: 10.3390/biology11091327 |
| [41] |
杨关福, 张细权, 李加琪, 聂龙, 王文. 徐闻黄牛和海南黄牛血液蛋白的遗传多样性. 华南农业大学学报, 1996, 17(2): 23-27.
|
|
|
| [1] | 寇淑君, 霍阿红, 付国庆, 纪军建, 王瑶, 左振兴, 刘敏轩, 陆平. 利用荧光SSR分析中国糜子的遗传多样性和群体遗传结构[J]. 中国农业科学, 2019, 52(9): 1475-1477. |
| [2] | 魏潇,章秋平,刘宁,张玉萍,徐铭,刘硕,张玉君,马小雪,刘威生. 不同来源中国李(Prunus salicina L.)的多样性与近缘种关系[J]. 中国农业科学, 2019, 52(3): 568-578. |
| [3] | 赵久然,李春辉,宋伟,王元东,张如养,王继东,王凤格,田红丽,王蕊. 基于SNP芯片揭示中国玉米育种种质的遗传多样性与群体遗传结构[J]. 中国农业科学, 2018, 51(4): 626-634. |
| [4] | 董合忠,张艳军,张冬梅,代建龙,张旺锋. 基于集中收获的新型棉花群体结构[J]. 中国农业科学, 2018, 51(24): 4615-4624. |
| [5] | 薛延桃,陆平,乔治军,刘敏轩,王瑞云. 基于SSR标记的黍稷种质资源遗传多样性及亲缘关系研究 [J]. 中国农业科学, 2018, 51(15): 2846-2859. |
| [6] | 时佳,翟胜男,刘金栋,魏景欣,白璐,高文伟,闻伟锷,何中虎,夏先春,耿洪伟. 普通小麦籽粒过氧化物酶活性全基因组关联分析[J]. 中国农业科学, 2017, 50(21): 4212-4227. |
| [7] | 曹亦兵,黄收兵,王媛媛,夏雨晴,孟庆锋,陶洪斌,王璞. 玉米群体生长与光截获的动态模拟及应用[J]. 中国农业科学, 2017, 50(11): 1973-1981. |
| [8] | 罗凯,卢会翔,吴正丹,吴雪莉,尹旺,唐道彬,王季春,张凯. 中国西南地区甘薯主要育种亲本的遗传多样性及群体结构分析[J]. 中国农业科学, 2016, 49(3): 593-608. |
| [9] | 张佳蕾,郭峰,杨佃卿,孟静静,杨莎,王兴语,陶寿祥,李新国,万书波. 单粒精播对超高产花生群体结构和产量的影响[J]. 中国农业科学, 2015, 48(18): 3757-3766. |
| [10] | 聂新辉,尤春源,鲍健,李晓方,惠慧,刘洪亮,秦江鸿,林忠旭. 基于关联分析的新陆早棉花品种农艺和纤维品质性状优异等位基因挖掘[J]. 中国农业科学, 2015, 48(15): 2891-2910. |
| [11] | 谭龙涛,喻春明,陈平,王延周,陈继康,温岚,郑建树,熊和平. 苎麻自交纯合进度研究[J]. 中国农业科学, 2014, 47(22): 4371-4379. |
| [12] | 强海平1, 余国辉1, 刘海泉1, 高洪文1, 刘贵波2, 赵海明2, 王赞1. 基于SSR标记的中美紫花苜蓿品种遗传多样性研究[J]. 中国农业科学, 2014, 47(14): 2853-2862. |
| [13] | 章秋平, 刘冬成, 刘威生, 刘硕, 张爱民, 刘宁, 张玉萍. 华北生态群普通杏遗传多样性与群体结构分析[J]. 中国农业科学, 2013, 46(1): 89-98. |
| [14] | 范虎, 赵团结, 丁艳来, 邢光南, 盖钧镒. 中国野生大豆群体特征和地理分化的遗传分析[J]. 中国农业科学, 2012, 45(3): 414-425. |
| [15] | 刘志斋, 吴迅, 刘海利, 李永祥, 李清超, 王凤格, 石云素, 宋燕春, 宋伟彬, 赵久然, 赖锦盛, 黎裕, 王天宇. 基于40个核心SSR标记揭示的820份中国玉米重要自交系的遗传多样性与群体结构[J]. 中国农业科学, 2012, 45(11): 2107-2138. |
|
||