













中国农业科学 ›› 2022, Vol. 55 ›› Issue (3): 503-513.doi: 10.3864/j.issn.0578-1752.2022.03.007
收稿日期:2021-07-14
接受日期:2021-08-26
出版日期:2022-02-01
发布日期:2022-02-11
通讯作者:
黄丽丽
作者简介:张晋龙,E-mail: 基金资助:
ZHANG JinLong( ),ZHAO ZhiBo,LIU Wei,HUANG LiLi(
),ZHAO ZhiBo,LIU Wei,HUANG LiLi( )
)
Received:2021-07-14
Accepted:2021-08-26
Online:2022-02-01
Published:2022-02-11
Contact:
LiLi HUANG
摘要: 目的 由丁香假单胞菌猕猴桃致病变种(Pseudomonas syringae pv. actinidiae,Psa)引起的猕猴桃细菌性溃疡病是全球猕猴桃产业最具毁灭性的病害。病原细菌主要通过III型分泌系统(type III secretion system,T3SS)将多种效应蛋白(T3SS effector,T3SE)注入寄主植物细胞,进而促进病菌侵染和致病。本研究旨在解析Psa基因组中T3SE的信息并对其T3SS和T3SE的致病功能进行系统分析,为溃疡病菌致病机制的研究和防治策略的制定提供依据。方法 利用marker-free同源重组基因敲除技术获得M228菌株的T3SS功能缺陷突变体ΔhrcS和ΔhrcC,观察突变体在寄主上的致病力,同时检测突变体诱导本氏烟产生细胞坏死的情况;随后利用从Pseudomonas-Plant Interaction数据库下载的T3SE数据库,本地BLAST多序列比对构建强、弱致病菌株M228和M227的T3SE库,并对二者的T3SE基因信息进行比对分析;另外,获得M228菌株T3SE单、多效应子突变菌株20株及2株HopR1基因回补菌株(共涉及19个T3SE),并将各突变体室内有伤接菌猕猴桃枝条,系统评价各突变体致病力变化并进行统计分析。结果 通过对Psa的hrcS和hrcC基因进行突变,证明T3SS是其在寄主上致病以及非寄主上过敏性坏死反应(HR)所必需的。通过数据库同源比对,发现在强毒株系和弱毒株系中有31个T3SE基因具有100%的同源性,选取一些基因进行缺失突变,发现hopM1/avrE1和hopR1是Psa重要的毒性因子,且二者不存在功能冗余。另外,单独敲除avrPto5或avrRpm1均能提高Psa致病力。在缺失A-F-E基因簇和avrPto5的菌株中,敲除hopM1/avrE1和hopR1也分别导致Psa的致病力显著下降;而同时敲除hopM1/avrE1、hopR1、avrPto5和A-F-E基因簇导致病菌完全丧失致病力。结论HopM1/AvrE1与同家族HopR1均为Psa重要致病因子,且独立于其他效应子发挥作用;avrPto5和avrRpm1基因缺失可以增强Psa的致病力。
张晋龙,赵志博,刘巍,黄丽丽. 猕猴桃细菌性溃疡病菌T3SS关键效应蛋白基因致病功能[J]. 中国农业科学, 2022, 55(3): 503-513.
ZHANG JinLong,ZHAO ZhiBo,LIU Wei,HUANG LiLi. The Function of Key T3SS Effectors in Pseudomonas syringae pv. actinidiae[J]. Scientia Agricultura Sinica, 2022, 55(3): 503-513.
表1
敲除和回补突变体信息汇总"
| 突变体名称 The name of mutants | 详细信息 Related detail information |
|---|---|
| ΔhrcC | T3SS结构基因hrcC功能缺陷突变体 hrcC-deletion generating the T3SS-deficient mutant |
| ΔhrcS | T3SS结构基因hrcS功能缺陷突变体hrcS-deletion generating the T3SS-deficient mutant |
| ΔO | T3SE hopM1/avrE1缺失突变体T3SE hopM1/avrE1-deletion mutant |
| ΔM | T3SE hopR1缺失突变体T3SE hopR1-deletion mutant |
| ΔOM | T3SE hopM1/avrE1+hopR1缺失突变体T3SE hopR1-deletion mutant based on ΔO background |
| ΔMO | T3SE hopR1+hopM1/avrE1缺失突变体T3SE hopM1/avrE1-deletion mutant based on ΔM background |
| ΔM-C-hopR1 | T3SE hopR1突变体基础上回补hopR1的回补菌株T3SE hopR1-complement mutant based on ΔM background |
| ΔOM-C-hopR1 | T3SE hopR1+hopM1/AvrE1突变体基础上回补hopR1的回补菌株T3SE hopR1-complement mutant based on ΔOM background |
| ΔA | |
| ΔF | |
| ΔE | |
| ΔAF | |
| ΔAFE | |
| ΔAFEL | 在ΔAFE突变体的背景下敲除avrPto5获得突变体avrPto5-deletion mutant based on ΔAFE background |
| ΔAFELM | 在ΔAFEL突变体的背景下敲除hopR1获得突变体hopR1-deletion mutant based on ΔAFEL background |
| ΔAFELO | 在ΔAFEL突变体的背景下敲除hopM1/avrE1获得突变体hopM1/avrE1-deletion mutant based on ΔAFEL background |
| ΔAFELOM | 在ΔAFEL突变体的背景下同时敲除hopM1/avrE1和hopR1获得突变体 hopM1/avrE1- and hopR1-deletion mutant based on ΔAFEL background |
| ΔOMN | 在ΔOM突变体基础上敲除avrRpm1获得突变体avrRpm1-deletion mutant based on ΔOM background |
表2
Psa M228和M227菌株的T3SE库"
| 位置 Locus | T3SE | ICMP 18884 | 伴侣分子 Chaperon | M228 | M227 | 基因敲除标识符 Symbol of gene deletion | ||
|---|---|---|---|---|---|---|---|---|
| HopY1 | IYO_000845 | |||||||
| Cluster A | HopQ1 | IYO_003525 | A | |||||
| HopD1 | IYO_003530 | |||||||
| AvrD1 | IYO_003570 | |||||||
| AvrB4-l | IYO_003600 | |||||||
| HopAYl | IYO_003610 | |||||||
| HopX3 | IYO_003635 | ShcF | ||||||
| HopAW1 | IYO_003657 | |||||||
| HopBB1-2 | IYO_003675 | ShcF | ||||||
| HopAF1 | IYO_003680 | |||||||
| HopAO2 | IYO_003720 | |||||||
| HopBB1-1 | IYO_003727 | ShcF | ||||||
| HopS2 | IYO_004052 | ShcS2 | ||||||
| HopI1 | IYO_005160 | |||||||
| CEL/Cluster O | HopN1 | IYO_006735 | ShcN | |||||
| HopAA1-1 | IYO_006745 | |||||||
| HopM1 | IYO_006760 | ShcM | O | |||||
| AvrE1 | IYO_006770 | ShcE | ||||||
| AvrRpm1 | IYO_008065 | N | ||||||
| Cluster E | HopZ5 | IYO_008282 | E | |||||
| HopH1 | IYO_008285 | |||||||
| HopAM1-2 | IYO_008385 | |||||||
| HopAE1 | IYO_012225 | |||||||
| HopAZ1 | IYO_018555 | |||||||
| AvrPto5 | IYO_020425 | L | ||||||
| HonAM1-1 | IYO_023205 | |||||||
| Cluster F | HopAI1 | IYO_023980 | F | |||||
| HopAH1 | IYO_023985 | |||||||
| HopAG1 | IYO_023990 | |||||||
| Cluster M | HopR1 | IYO_024150 | M | |||||
| HopW1 | IYO_024160 | |||||||
| HopF2 | IYO_024217 | ShcF | ||||||
| HopAS1 | IYO_027420 | |||||||
| HopA1 | IYO_028375 | ShcA | ||||||
| HopZ3 | IYO_029045 | Tir | ||||||
| AvrRpm2 | IYO_029288 | ShcF | ||||||
| Plasmid | HopAV1 | IYO_029710 | ||||||
| HopAA1-2 | IYO_029780-795 | |||||||
| HopAU1 | IYO_029795 | |||||||
| 存在的基因 Present | 差异的基因 Variable | 不完整的基因 Incomplete | ||||||

图1
Psa的T3SS缺陷突变体不能致病,不能诱导烟草产生HRa:Psa菌株M228及其ΔhrcC、ΔhrcS 突变体接种‘红阳’猕猴桃枝条,2×108 cfu/mL(15 dpi) Psa M228 strain and its T3SS-deficient mutants ΔhrcC, ΔhrcS were inoculated on canes of ‘HongYang’ with 2×108 cfu/mL concentration (15 days post inoculation);b:接种叶盘,上排为‘红阳’猕猴桃叶片,下排为‘翠香’猕猴桃叶片,104 cfu/mL(5 dpi)Leaf discs of ‘HongYang’ (upper) and ‘CuiXiang’ (bottom) with 104 cfu/mL concentration (5 days post inoculation);c:注射本氏烟叶片,108 cfu/mL(2 dpi)N. benthamina leaves with 108 cfu/mL concentration (2 days post inoculation)。每菌株接种至少10个枝条或叶盘,每菌株至少注射3株烟草的3个叶片。试验重复至少2次,得相似结果 For the inoculation, at least 10 canes or leaf discs were used for each strain, and at least three tobacco leaves from three plants were treated with each strain. Experiments were repeated at least twice with similar results"
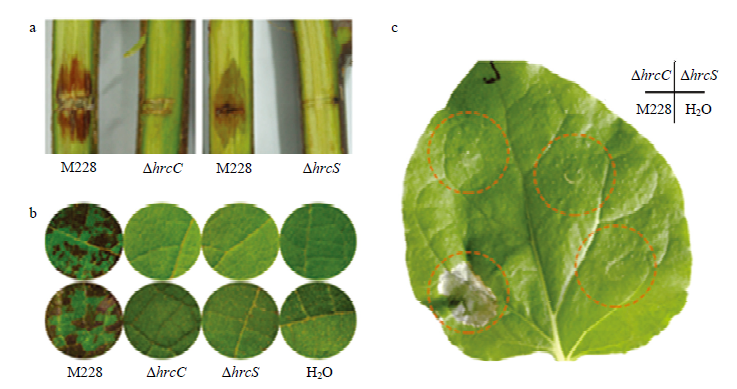

图3
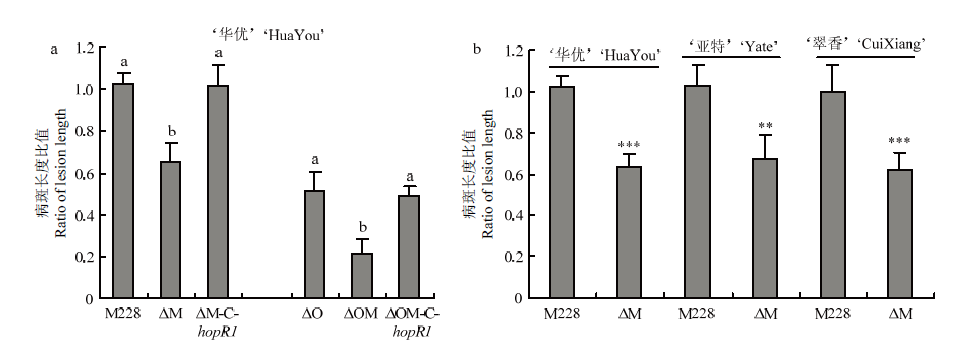
敲除hopR1导致致病力显著下降a:接种至‘华优’Inoculation on cultivar ‘HuaYou’;b:野生型菌株M228及其hopR1突变体分别接种至‘亚特’‘翠香’和‘华优’M228 and the hopR1 mutant were inoculated on ‘Yate’ ‘CuiXiang’ and ‘HuaYou’。试验至少重复2次,得相似结果Experiments were repeated at least twice with similar results (Student’s t-test or Duncan’s multiple range test, P<0.01)"
| [1] | 秦虎强, 高小宁, 赵志博, 朱穗层, 李建民, 黄丽丽. 陕西猕猴桃细菌性溃疡病田间发生动态和规律. 植物保护学报, 2013, 40(3): 225-230. |
| QIN H Q, GAO X N, ZHAO Z B, ZHU H C, LI J M, HUANG L L.The prevalence dynamics and rules of bacterial canker of kiwifruit in Shaanxi. Acta Phytophylacica Sinica, 2013, 40(3): 225-230. (in Chinese) | |
| [2] | 高小宁, 赵志博, 黄其玲, 秦虎强, 黄丽丽. 猕猴桃细菌性溃疡病研究进展. 果树学报, 2012, 29(2): 262-268. |
| GAO X N, ZHAO Z B, HUANG Q L, QIN H Q, HUANG L L.Advances in research on bacterial canker of kiwifruit. Journal of Fruit Science, 2012, 29(2): 262-268. (in Chinese) | |
| [3] |
ZHAO Z, CHEN J, GAO X, ZHANG D, ZHANG J, WEN J, QIN H, GUO M, HUANG L.Comparative genomics reveal pathogenicity- related loci in Pseudomonas syringae pv. actinidiae biovar 3. Molecular Plant Pathology, 2019, 20(7): 923-942.
doi: 10.1111/mpp.2019.20.issue-7 |
| [4] | 孙思, 牛建军, 王岱. 细菌三型分泌系统效应蛋白转运的研究进展. 微生物学报, 2017, 57(10): 1452-1460. |
| SUN S, NIU J J, WANG D.Advances in studies of translocation of effector by bacterial type 3 secretion system. Acta Microbiologica Sinica, 2017, 57(10): 1452-1460. (in Chinese) | |
| [5] |
LINDEBERG M, CUNNAC S, COLLMER A.Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends in Microbiology, 2012, 20(4): 199-208.
doi: 10.1016/j.tim.2012.01.003 |
| [6] |
DOS SANTOS A M P,FERRARI R G, CONTE-JUNIOR C A. Type three secretion system in Salmonella typhimurium: The key to infection. Genes and Genomics, 2020, 42(5): 495-506.
doi: 10.1007/s13258-020-00918-8 |
| [7] | 朱秀秀, 高必达, 赵廷昌, 张月娟. 植物病原细菌Ⅲ型分泌系统及Pseudomonas syringae pv. tomato的信号分子分泌研究进展. 湖南农业科学, 2009(2): 19-22. |
| ZHU X X, GAO B D, ZHAO Y C, ZHANG Y J.Research progress of type III secretory system of plant pathogenic bacteria and signal molecule secretion of Pseudomonas syringae pv. tomato. Hunan Agricultural Sciences, 2009(2): 19-22. (in Chinese) | |
| [8] |
MARLOVITS T C, KUBORI T, LARA-TEJERO M, THOMAS D, UNGER V M, GALAN J E.Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature, 2006, 441(7093): 637-640.
doi: 10.1038/nature04822 |
| [9] |
MACHO A P, ZIPFEL C.Plant PRRs and the activation of innate immune signaling. Molecular Cell, 2014, 54(2): 263-272.
doi: 10.1016/j.molcel.2014.03.028 |
| [10] | 温晶. 猕猴桃溃疡病菌Ⅲ型效应蛋白的筛选及效应蛋白HopX3功能的初步研究[D]. 杨凌: 西北农林科技大学, 2016. |
| WEN J.Identification of Psa type III effectors and preliminary analysis of effector HopX3 in pathogenicity[D]. Yangling: Northwest A&F University, 2016. (in Chinese) | |
| [11] | BALTRUS D A, NISHIMURA M T, ROMANCHUK A, CHANG J H,MUKHTAR M S, CHERKIS K, ROACH J, GRANT S R, JONES C D, DANGL J L.Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathogens, 2011, 7(7): e1002132. |
| [12] | CUNNAC S, CHAKRAVARTHY S, KVITKO B H, RUSSELL A B, MARTIN G B, COLLMER A.Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(7): 2975-2980. |
| [13] |
KVITKO B H, PARK D H, VELASQUEZ A C, WEI C F, RUSSELL A B, MARTIN G B, SCHNEIDER D J, COLLMER A.Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathogens, 2009, 5(4): e1000388.
doi: 10.1371/journal.ppat.1000388 |
| [14] |
TAMPAKAKI A P, SKANDILIS N, GAZI A D, BASTAKI M N, PANAGIOTIS F S, CHAROVA S N, KOKKINIDIS M, PANOPOULOS N J.Playing the “Harp”: Evolution of our understanding of hrp/hrc genes 1. Annual Review of Phytopathology, 2010, 48: 347-370.
doi: 10.1146/phyto.2010.48.issue-1 |
| [15] |
CHOI S, JAYARAMAN J, SEGONZAC C, PARK H J, PARK H, HAN S W, SOHN K H.Pseudomonas syringae pv. actinidiae type III effectors localized at multiple cellular compartments activate or suppress innate immune responses in Nicotiana benthamiana. Frontiers in Plant Science, 2017, 8: 2157.
doi: 10.3389/fpls.2017.02157 |
| [16] |
JAYARAMAN J, CHOI S, PROKCHORCHIK M, CHOI D S, SPIANDORE A, RIKKERINK E H, TEMPLETON M D, SEGONZAC C, SOHN K H.A bacterial acetyltransferase triggers immunity in Arabidopsis thaliana independent of hypersensitive response. Scientific Reports, 2017, 7(1): 3557.
doi: 10.1038/s41598-017-03704-x |
| [17] |
CHOI S, JAYARAMAN J, SOHN K H.Arabidopsis thaliana SOBER1 (SUPPRESSOR OF AVRBST-ELICITED RESISTANCE 1) suppresses plant immunity triggered by multiple bacterial acetyltransferase effectors. New Phytologist, 2018, 219(1): 324-335.
doi: 10.1111/nph.2018.219.issue-1 |
| [18] | YOON M, RIKKERINK E H A. Rpa1 mediates an immune response to avrRpm1Psa and confers resistance against Pseudomonas syringae pv. actinidiae. The Plant Journal, 2020, 102(4): 688-702. |
| [19] |
JAYARAMAN J, YOON M, APPLEGATE E R, STROUD E A, TEMPLETON M D.AvrE1 and HopR1 from Pseudomonas syringae pv. actinidiae are additively required for full virulence on kiwifruit. Molecular Plant Pathology, 2020, 21(11): 1467-1480.
doi: 10.1111/mpp.12989 |
| [20] | 赵志博. 猕猴桃细菌性溃疡病菌群体结构与致病机制研究[D]. 杨凌: 西北农林科技大学, 2016. |
| ZHAO Z B.Population composition and pathogenetic mechanism in Psuesdomonas syringae pv. actinidiae[D]. Yangling: Northwest A&F University, 2016. (in Chinese) | |
| [21] |
WANG K, KANG L, ANAND A, LAZAROVITS G, MYSORE K S.Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytologist, 2007, 174(1): 212-223.
doi: 10.1111/nph.2007.174.issue-1 |
| [22] | KVITKO B H, COLLMER A.Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains//Plant Immunity. Methods and Protocols, 2011, 712: 109-128. |
| [23] |
SAWADA H, FUJIKAWA T.Genetic diversity of Pseudomonas syringae pv. actinidiae, pathogen of kiwifruit bacterial canker. Plant Pathology, 2019, 68(7): 1235-1248.
doi: 10.1111/ppa.v68.7 |
| [24] |
XIN X F, NOMURA K, AUNG K, VELASQUEZ A C, YAO J, BOUTROT F, CHANG J H, ZIPFEL C, HE S Y.Bacteria establish an aqueous living space in plants crucial for virulence. Nature, 2016, 539(7630): 524-529.
doi: 10.1038/nature20166 |
| [25] |
JIN L, HAM J H, HAGE R, ZHAO W, SOTO-HERNANDEZ J, LEE S Y, PAEK S M, KIM M G, BOONE C, COPLIN D L, MACKEY D.Direct and indirect targeting of PP2A by conserved bacterial type-III effector proteins. PLoS Pathogens, 2016, 12(5): e1005609.
doi: 10.1371/journal.ppat.1005609 |
| [26] |
DEGRAVE A, SIAMER S, BOUREAU T, BARNY M A.The AvrE superfamily: Ancestral type III effectors involved in suppression of pathogen-associated molecular pattern-triggered immunity. Molecular Plant Pathology, 2015, 16(8): 899-905.
doi: 10.1111/mpp.2015.16.issue-8 |
| [27] | PALACE S G, PROULX M K, SZABADY R L, GOGUEN J D.Gain-of-function analysis reveals important virulence roles for the Yersinia pestis type III secretion system effectors YopJ, YopT, and YpkA. Infection and Immunity, 2018, 86(9): e00318-18. |
| [28] |
ÜSTÜN S, KÖNIG P, GUTTMAN D S, BÖRNKE F. HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Molecular Plant-Microbe Interactions, 2014, 27(7): 611-623.
doi: 10.1094/MPMI-12-13-0363-R |
| [29] | LEWIS J D, LEE A H Y,HASSAN J A,WAN J,HURLEY B,JHINGREE J R,WANG P W,LO T,YOUN J Y,GUTTMAN D S,DESVEAUX D. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(46): 18722-18727. |
| [30] |
LEWIS J D, LEE A, MA W B, ZHOU H B, GUTTMAN D S, DESVEAUX D.The YopJ superfamily in plant-associated bacteria. Molecular Plant Pathology, 2011, 12(9): 928-937.
doi: 10.1111/j.1364-3703.2011.00719.x |
| [31] |
ZHOU H B, LIN J A, JOHNSON A, MORGAN R L, ZHONG W W, MA W B.Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host and Microbe, 2011, 9(3): 177-186.
doi: 10.1016/j.chom.2011.02.007 |
| [32] |
MACHO A P, GUIDOT A, BARBERIS P, BEUZON C R, GENIN S.A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Molecular Plant-Microbe Interactions, 2010, 23(9): 1197-1205.
doi: 10.1094/MPMI-23-9-1197 |
| [33] |
BARTETZKO V, SONNEWALD S, VOGEL F, HARTNER K, STADLER R, HAMMES U Z, BORNKE F.The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: Evidence for interference with cell wall-associated defense eesponses. Molecular Plant-Microbe Interactions, 2009, 22(6): 655-664.
doi: 10.1094/MPMI-22-6-0655 |
| [34] |
CHEN H, HU Y, QIN K Y, YANG X Z, JIA Z J, LI Q, CHEN H B, YANG H.A serological approach for the identification of the effector hopz5 of Pseudomonas syringae pv. actinidiae: A tool for the rapid immunodetection of kiwifruit bacterial canker. Journal of Plant Pathology, 2018, 100(2): 171-177.
doi: 10.1007/s42161-018-0041-y |
| [35] |
KRAUS C M, MUNKVOLD K R, MARTIN G B.Natural variation in tomato reveals differences in the recognition of AvrPto and AvrPtoB effectors from Pseudomonas syringae. Molecular Plant, 2016, 9(5): 639-649.
doi: 10.1016/j.molp.2016.03.001 |
| [36] |
KIM M G, GENG X, LEE S Y, MACKEY D.The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. The Plant Journal, 2009, 57(4): 645-653.
doi: 10.1111/tpj.2009.57.issue-4 |
| [1] | 黄家权,李莉,吴丰年,郑正,邓晓玲. 携带不同原噬菌体的黄龙病菌在柑橘木虱体内的增殖及致病力[J]. 中国农业科学, 2022, 55(4): 719-728. |
| [2] | 张承启,廖露露,齐永霞,丁克坚,陈莉. 禾谷镰孢核孔蛋白基因FgNup42的功能分析[J]. 中国农业科学, 2021, 54(9): 1894-1903. |
| [3] | 曹钰晗,李紫腾,张静怡,张静娜,胡同乐,王树桐,王亚南,曹克强. 我国苹果斑点落叶病菌携带dsRNA分析及一种dsRNA病毒的鉴定[J]. 中国农业科学, 2021, 54(22): 4787-4799. |
| [4] | 赵静雅,夏荟清,彭梦雅,凡卓,殷悦,徐赛博,张楠,陈文波,陈琳琳. 假禾谷镰孢转录因子FpAPSES的鉴定与功能分析[J]. 中国农业科学, 2021, 54(16): 3428-3439. |
| [5] | 王宝宝,郭成,孙素丽,夏玉生,朱振东,段灿星. 玉米穗腐病致病禾谷镰孢复合种的遗传多样性、致病力与毒素化学型分析[J]. 中国农业科学, 2020, 53(23): 4777-4790. |
| [6] | 孙琦,何芳,邵胜楠,刘政,黄家风. 棉花黄萎病菌VdHP1的克隆及功能分析[J]. 中国农业科学, 2020, 53(14): 2872-2884. |
| [7] | 齐悦,吕峻元,张悦,韦杰,张娜,杨文香,刘大群. 小麦叶锈菌效应蛋白Pt18906激发TcLr27+31的双层防御反应[J]. 中国农业科学, 2020, 53(12): 2371-2384. |
| [8] | 彭军波,李兴红,张玮,周莹,黄金宝,燕继晔. 葡萄溃疡病菌外泌蛋白LtGH61A的致病力及基因表达模式[J]. 中国农业科学, 2019, 52(24): 4518-4526. |
| [9] | 张一豪,冯鸿杰,袁媛,靳羽莹,师勇强,张朝军,李付广. 大丽轮枝菌弱致病力菌株Vd171对棉花黄萎病的诱导免疫作用及机制[J]. 中国农业科学, 2018, 51(6): 1067-1078. |
| [10] | 孙骏翔,张乾义,徐和敏,王团结,徐璐,邹兴启,朱元源,李翠,夏应菊,徐嫄,陈锴,张玉杰,赵启祖,王琴. 猪瘟病毒中等致病力毒株在体内的动态分布[J]. 中国农业科学, 2018, 51(21): 4146-4156. |
| [11] | 尹馨,马树杰,李梅,邓国华,侯玉杰,崔鹏飞,施建忠,陈化兰. PB2蛋白E627V突变可增强H7N9病毒对小鼠的致病力[J]. 中国农业科学, 2018, 51(17): 3379-3388. |
| [12] | 张俊祥,冀志蕊,王娜,徐成楠,迟福梅,周宗山. 苹果炭疽叶枯病菌GcAP1复合体β亚基基因的克隆及功能分析[J]. 中国农业科学, 2017, 50(8): 1430-1439. |
| [13] | 薄惠文,俞咪娜,于俊杰,尹小乐,丁慧,王亚会,刘永锋. 稻曲病菌T-DNA插入突变菌株B1241侧翼基因克隆[J]. 中国农业科学, 2016, 49(9): 1685-1695. |
| [14] | 许春景,吴玉星,戴青青,李正鹏,高小宁,黄丽丽. 苹果树腐烂病菌多聚半乳糖醛酸酶基因Vmpg7和Vmpg8的功能[J]. 中国农业科学, 2016, 49(8): 1489-1498. |
| [15] | 王宇秋,李国邦,杨娟,黎良,赵志学,樊晶,王文明. 稻曲病菌侵染水稻颖花的酵母双杂交cDNA文库构建与应用[J]. 中国农业科学, 2016, 49(5): 865-873. |
|
||