













中国农业科学 ›› 2025, Vol. 58 ›› Issue (15): 2960-2979.doi: 10.3864/j.issn.0578-1752.2025.15.003
庄润杰1,2( ), 刘慧铭1,2, 王诗雨1,2, 吕婉萍1,2, 温永仙1,2,*(
), 刘慧铭1,2, 王诗雨1,2, 吕婉萍1,2, 温永仙1,2,*( )
)
收稿日期:2025-02-07
接受日期:2025-05-16
出版日期:2025-08-01
发布日期:2025-07-30
通信作者:
联系方式:
庄润杰,E-mail:2215372440@qq.com。
基金资助:
ZHUANG RunJie1,2( ), LIU HuiMing1,2, WANG ShiYu1,2, LÜ WanPing1,2, WEN YongXian1,2,*(
), LIU HuiMing1,2, WANG ShiYu1,2, LÜ WanPing1,2, WEN YongXian1,2,*( )
)
Received:2025-02-07
Accepted:2025-05-16
Published:2025-08-01
Online:2025-07-30
摘要:
【目的】 全基因组选择(genomic selection,GS)是一种通过全基因组标记信息预测个体表型或遗传值的核心技术,在农业育种和遗传研究中具有重要的理论价值和实践意义。然而,高维特征冗余和非线性关系建模是全基因组选择中的关键挑战。提出一种从基因型到表型的堆叠集成模型(genotype to phenotype stacking ensemble,G2PSE),旨在提高预测精度和泛化能力,为高维基因组数据分析提供高效的解决方案。【方法】 构建G2PSE堆叠集成模型框架,综合应用十折交叉验证、集成学习、特征选择(LAR算法)和特征增强策略。模型采用随机森林(RF)、支持向量回归(SVR)和梯度提升回归(GBR)作为基学习器,并以普通最小二乘回归(OLSR)作为元学习器,同时,评估随机森林、支持向量回归和神经网络等元学习器对模型性能的影响。G2PSE模型包含3种核心子模型:(1)全特征堆叠集成(AFSE),充分利用所有SNP特征;(2)LAR特征堆叠集成(LFSE),通过特征选择减少冗余信息,提升泛化能力;(3)LAR特征增强堆叠集成(LFESE),结合特征选择与增强策略,在高维数据环境中优化预测能力。探讨3种特征增强变体(AFESE、HFESEⅠ、HFESEⅡ)的性能。最终,模型在小麦、大豆、罗非鱼3个物种的多性状数据集上进行试验评估,并进一步利用Pepper203数据集进行独立测试集评估,验证模型的鲁棒性。【结果】 G2PSE模型在皮尔逊相关系数(PCC)和平均绝对误差(MAE)2项指标上显著优于传统方法和单一机器学习模型。3种核心子模型中,LFESE通过结合特征选择与增强策略表现最佳,LFSE通过特征选择减少了冗余信息并增强了泛化能力,而AFSE在全面捕获基因型全局信息方面具有显著优势。此外,3种特征增强变体模型进一步验证了特征质量相较于特征数量对提升预测性能的重要性。试验还表明,在元学习器选择中,线性回归模型的表现最佳,而在计算效率上,LFESE和LFSE子模型展示了较为均衡的性能。且合理的特征选择阈值对模型性能至关重要,其中,低维数据集的最优阈值为10%—20%,而高维数据集的最优阈值为1%。最后,在独立测试集上的评估证明LFESE子模型具有最佳的泛化能力。【结论】 G2PSE模型通过集成学习、特征选择与增强策略显著提升了全基因组选择的预测性能。
庄润杰, 刘慧铭, 王诗雨, 吕婉萍, 温永仙. 基于G2PSE堆叠集成的全基因组选择方法[J]. 中国农业科学, 2025, 58(15): 2960-2979.
ZHUANG RunJie, LIU HuiMing, WANG ShiYu, LÜ WanPing, WEN YongXian. Genomic Selection Method Based on G2PSE Stacking Ensemble[J]. Scientia Agricultura Sinica, 2025, 58(15): 2960-2979.
表1
试验数据集相关信息"
| 数据集 Datasets | 性状 Traits | 个体数 Individuals | 标记数 SNPs | 遗传率 h2 | 数据来源 Data source |
|---|---|---|---|---|---|
| Wheat599 | E1-GY | 599 | 1279 | 0.832 | https://github.com/AIBreeding/DNNGP?tab=readme-ov-file |
| E2-GY | 599 | 1279 | 0.729 | ||
| E3-GY | 599 | 1279 | 0.689 | ||
| E4-GY | 599 | 1279 | 0.711 | ||
| Wheat2000 | TKW | 2000 | 33709 | 0.833 | https://github.com/cma2015/DeepGS |
| TW | 2000 | 33709 | 0.754 | ||
| GL | 2000 | 33709 | 0.881 | ||
| GW | 2000 | 33709 | 0.848 | ||
| GH | 2000 | 33709 | 0.839 | ||
| GP | 2000 | 33709 | 0.625 | ||
| SDS | 2000 | 33709 | 0.681 | ||
| PHT | 2000 | 33709 | 0.434 | ||
| Soy5014 | HT | 5014 | 4234 | 0.449 | https://doi.org/10.1534/g3.116.032268 |
| R8 | 5014 | 4234 | 0.558 | ||
| YLD | 5014 | 4234 | 0.485 | ||
| Tilapia1125 | HW | 1125 | 32306 | 0.304 | https://figshare.com/s/9b265a22b7e138c5a839 |
| Pepper203 | PHT | 203 | 14922 | 0.610 | https://bmcgenomdata.biomedcentral.com/articles/10.1186/s12863-023-01179-6 |
| FT | 203 | 14922 | 0.730 |
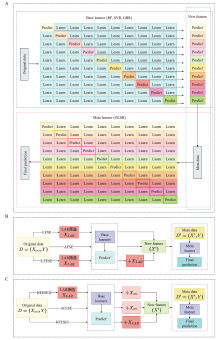
图1
G2PSE模型的总体架构及子模型设计示意图 A:G2PSE基础模型(AFSE)的训练与预测流程图,采用十折交叉验证方式训练3个基学习器(RF、SVR、GBR),其预测输出作为元学习器(OLSR)的输入特征,生成最终预测值;B:G2PSE的3个核心子模型框架,包括AFSE、LFSE、LFESE;C:G2PSE模型的3种特征增强变体框架,分别为AFESE、HFESEⅠ和HFESEⅡ;Xori:原始SNP特征,XLAR:通过LAR方法筛选的关键特征子集,X′:输入元学习器的特征(新特征),Y:表型值,D:原始数据集,D′:元数据集,Base learners:基学习器,Meta learner:元学习器,Predict′:基学习器的预测输出,Final prediction:最终预测输出"
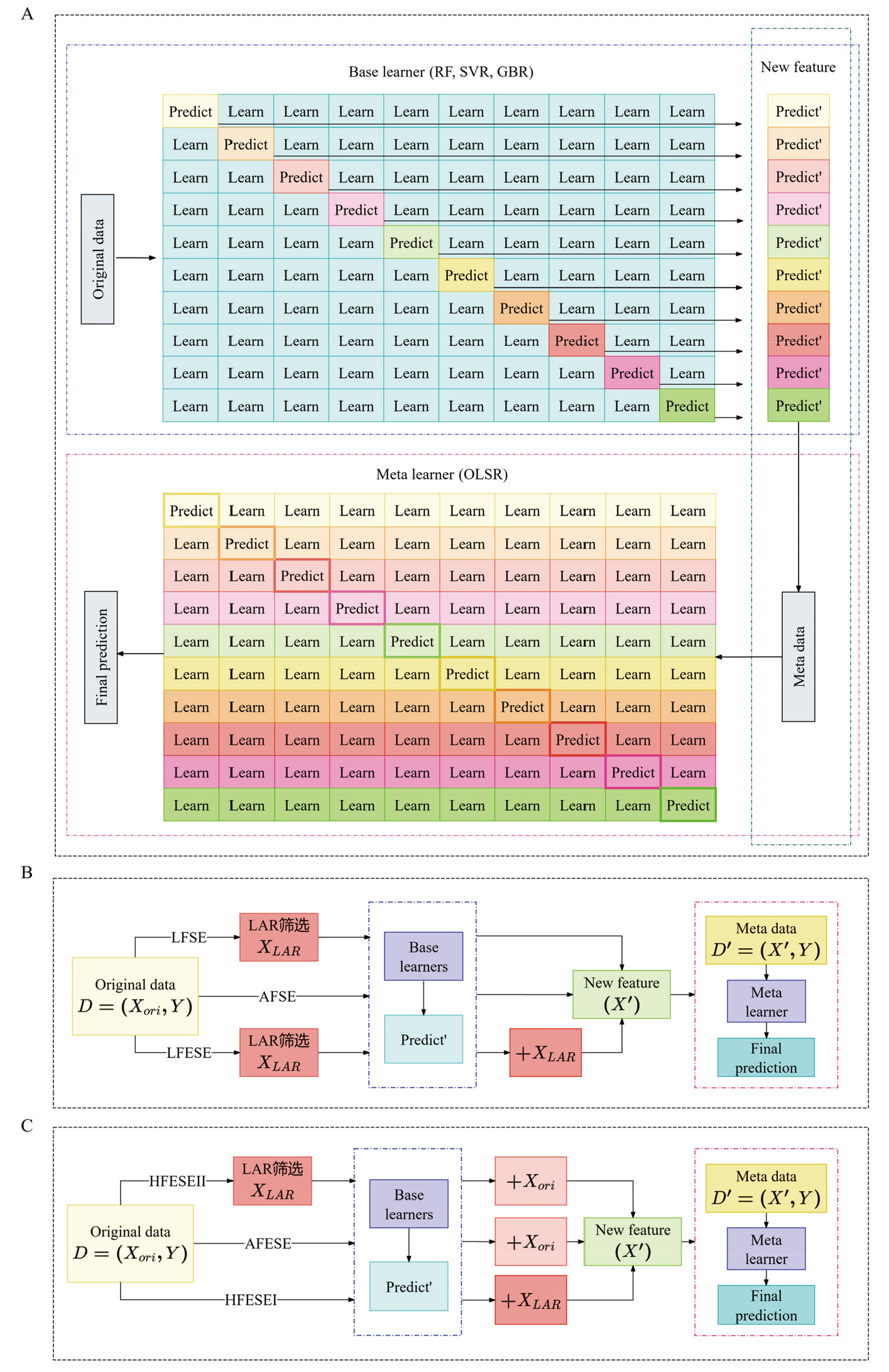
表2
Soy5014上G2PSE模型多重共线性检验"
| 性状Traits | 模型Model | 条件数Condition number |
|---|---|---|
| HT | AFSE | 14.823 |
| LFSE | 16.598 | |
| LFESE | 490.901 | |
| AFESE | 5.436×1016 | |
| HFESEⅠ | 298.676 | |
| HFESEⅡ | 5.281×1016 | |
| R8 | AFSE | 7.709 |
| LFSE | 8.837 | |
| LFESE | 495.610 | |
| AFESE | 5.506×1016 | |
| HFESEⅠ | 372.634 | |
| HFESEⅡ | 5.132×1016 | |
| YLD | AFSE | 8.353 |
| LFSE | 8.817 | |
| LFESE | 286.771 | |
| AFESE | 5.291×1016 | |
| HFESEⅠ | 286.676 | |
| HFESEⅡ | 5.489×1016 |
表3
Wheat599上不同LAR筛选阈值对于G2PSE模型预测性能的影响"
| 环境 Environment | 筛选SNP数目 Number of selected SNPs | LFSE模型 LFSE model | LFESE模型 LFESE model | HFESEⅠ模型 HFESEⅠ model | HFESEⅡ模型 HFESEⅡ model |
|---|---|---|---|---|---|
| E1-GY | 13 | 0.511(0.691) | 0.489 (0.701) | 0.581 (0.641) | 0.297 (1.132) |
| 64 | 0.592 (0.634) | 0.612 (0.616) | 0.563 (0.655) | 0.305 (1.112) | |
| 128 | 0.604 (0.631) | 0.636 (0.606) | 0.543 (0.683) | 0.321 (1.095) | |
| 256 | 0.604 (0.625) | 0.542 (0.670) | 0.454 (1.251) | 0.330 (1.074) | |
| 512 | 0.597 (0.629) | 0.407 (1.631) | 0.333(1.728) | 0.354 (1.030) | |
| 767 | 0.596 (0.630) | 0.129 (3.452) | 0.079 (3.518) | 0.353 (1.033) | |
| E2-GY | 13 | 0.429 (0.701) | 0.467 (0.691) | 0.518 (0.658) | 0.339 (1.095) |
| 64 | 0.592 (0.624) | 0.621 (0.622) | 0.614 (0.625) | 0.360 (1.033) | |
| 128 | 0.607 (0.612) | 0.698 (0.588) | 0.681 (0.592) | 0.373 (1.003) | |
| 256 | 0.598 (0.622) | 0.662 (0.634) | 0.630 (0.662) | 0.388 (0.968) | |
| 512 | 0.564 (0.638) | 0.521 (1.014) | 0.409 (1.243) | 0.431 (0.909) | |
| 767 | 0.536 (0.648) | 0.287 (2.026) | 0.204 (2.484) | 0.489 (0.838) | |
| E3-GY | 13 | 0.512 (0.678) | 0.493 (0.681) | 0.455 (0.692) | 0.276 (1.070) |
| 64 | 0.512 (0.677) | 0.498 (0.679) | 0.501 (0.689) | 0.274 (1.072) | |
| 128 | 0.555 (0.655) | 0.594 (0.628) | 0.551 (0.672) | 0.287 (1.059) | |
| 256 | 0.511 (0.678) | 0.496 (0.682) | 0.523 (0.743) | 0.280 (1.061) | |
| 512 | 0.510 (0.681) | 0.499 (0.679) | 0.285 (1.394) | 0.282 (1.061) | |
| 767 | 0.510 (0.680) | 0.496 (0.680) | 0.132 (3.048) | 0.279 (1.065) | |
| E4-GY | 13 | 0.457 (0.711) | 0.477 (0.686) | 0.514 (0.668) | 0.291 (1.079) |
| 64 | 0.604 (0.630) | 0.602 (0.633) | 0.592 (0.644) | 0.311 (1.047) | |
| 128 | 0.639 (0.607) | 0.653 (0.598) | 0.648 (0.614) | 0.319 (1.037) | |
| 256 | 0.628 (0.607) | 0.708 (0.594) | 0.668 (0.637) | 0.313 (1.044) | |
| 512 | 0.586 (0.628) | 0.434 (1.123) | 0.403 (1.157) | 0.323 (1.029) | |
| 767 | 0.560 (0.641) | 0.243 (2.231) | 0.230 (2.387) | 0.340 (1.011) |
表4
Wheat2000上不同LAR筛选阈值对于G2PSE模型预测性能的影响"
| 性状 Traits | 筛选SNP数目 Number of selected SNPs | LFSE模型 LFSE model | LFESE模型 LFESE model | HFESEⅠ 模型 HFESEⅠ model | HFESEⅡ 模型 HFESEⅡ model |
|---|---|---|---|---|---|
| TKW | 337 | 0.713 (0.562) | 0.802 (0.467) | 0.710 (0.572) | 0.669 (0.589) |
| 1685 | 0.710 (0.560) | 0.750 (0.643) | 0.522 (0.974) | 0.667 (0.590) | |
| 3371 | 0.704 (0.567) | 0.694 (0.648) | 0.404 (1.099) | 0.666 (0.591) | |
| 6742 | 0.704 (0.568) | 0.689 (0.662) | 0.373 (1.163) | 0.666 (0.591) | |
| 13483 | 0.702 (0.569) | 0.691 (0.660) | 0.368 (1.171) | 0.666 (0.591) | |
| 20225 | 0.700 (0.571) | 0.724 (0.612) | 0.365 (1.176) | 0.666(0.591) | |
| TW | 337 | 0.674 (0.564) | 0.778 (0.476) | 0.709 (0.573) | 0.602 (0.613) |
| 1685 | 0.665 (0.572) | 0.655 (0.822) | 0.528 (0.966) | 0.600 (0.614) | |
| 3371 | 0.661 (0.572) | 0.635 (0.737) | 0.374 (1.170) | 0.599 (0.615) | |
| 6742 | 0.657 (0.577) | 0.627 (0.731) | 0.373 (1.162) | 0.599 (0.615) | |
| 13483 | 0.658 (0.576) | 0.627 (0.733) | 0.368 (1.170) | 0.599 (0.615) | |
| 20225 | 0.659 (0.576) | 0.628 (0.731) | 0.366 (1.175) | 0.600 (0.615) | |
| GL | 337 | 0.772 (0.478) | 0.839 (0.410) | 0.754 (0.510) | 0.741 (0.502) |
| 1685 | 0.765 (0.486) | 0.738 (0.648) | 0.522 (0.947) | 0.740 (0.504) | |
| 3371 | 0.763 (0.489) | 0.699 (0.654) | 0.478 (1.007) | 0.739 (0.504) | |
| 6742 | 0.762 (0.490) | 0.703 (0.643) | 0.445 (1.031) | 0.739 (0.504) | |
| 13483 | 0.764 (0.488) | 0.704 (0.642) | 0.442 (1.036) | 0.739 (0.504) | |
| 20225 | 0.761 (0.490) | 0.702 (0.644) | 0.440 (1.039) | 0.739 (0.504) | |
| GW | 337 | 0.750 (0.514) | 0.812 (0.451) | 0.712 (0.557) | 0.732 (0.526) |
| 1685 | 0.745 (0.518) | 0.743 (0.654) | 0.497 (1.010) | 0.731 (0.526) | |
| 3371 | 0.740 (0.524) | 0.746 (0.582) | 0.420 (1.086) | 0.731 (0.527) | |
| 6742 | 0.741 (0.523) | 0.716 (0.634) | 0.402 (1.136) | 0.731 (0.527) | |
| 13483 | 0.741 (0.523) | 0.714 (0.636) | 0.401 (1.137) | 0.731 (0.527) | |
| 20225 | 0.742 (0.523) | 0.716 (0.634) | 0.400 (1.137) | 0.731 (0.527) | |
| GH | 337 | 0.687 (0.567) | 0.787 (0.483) | 0.677 (0.586) | 0.682 (0.567) |
| 1685 | 0.686 (0.566) | 0.707 (0.705) | 0.463 (1.043) | 0.681 (0.568) | |
| 3371 | 0.691 (0.561) | 0.695 (0.654) | 0.370 (1.170) | 0.680 (0.569) | |
| 6742 | 0.684 (0.566) | 0.698 (0.645) | 0.395 (1.125) | 0.680 (0.569) | |
| 13483 | 0.686 (0.565) | 0.698 (0.645) | 0.392 (1.129) | 0.680 (0.569) | |
| 20225 | 0.689 (0.563) | 0.699 (0.644) | 0.391 (1.132) | 0.680 (0.569) | |
| GP | 337 | 0.626 (0.604) | 0.746 (0.512) | 0.627 (0.622) | 0.515 (0.667) |
| 1685 | 0.609 (0.618) | 0.662 (0.800) | 0.414 (1.175) | 0.513 (0.668) | |
| 3371 | 0.604 (0.619) | 0.616 (0.759) | 0.358 (1.167) | 0.512 (0.668) | |
| 6742 | 0.603 (0.619) | 0.617 (0.757) | 0.365 (1.165) | 0.512 (0.668) | |
| 13483 | 0.603 (0.619) | 0.618 (0.756) | 0.357 (1.177) | 0.512 (0.668) | |
| 20225 | 0.602 (0.621) | 0.615 (0.760) | 0.352 (1.185) | 0.512 (0.668) | |
| SDS | 337 | 0.663 (0.599) | 0.796 (0.477) | 0.694 (0.575) | 0.525 (0.694) |
| 1685 | 0.656 (0.607) | 0.763 (0.606) | 0.580 (0.862) | 0.523 (0.695) | |
| 3371 | 0.644 (0.621) | 0.714 (0.626) | 0.468 (0.979) | 0.520 (0.697) | |
| 6742 | 0.644 (0.623) | 0.713 (0.629) | 0.488 (0.948) | 0.518 (0.698) | |
| 13483 | 0.643 (0.622) | 0.712 (0.631) | 0.487 (0.949) | 0.521 (0.696) | |
| 20225 | 0.644 (0.621) | 0.707 (0.636) | 0.440 (1.025) | 0.520 (0.697) | |
| PHT | 337 | 0.501 (0.664) | 0.697 (0.566) | 0.553 (0.673) | 0.279 (0.755) |
| 1685 | 0.477 (0.672) | 0.626 (0.844) | 0.422 (1.131) | 0.278 (0.755) | |
| 3371 | 0.449 (0.682) | 0.520 (0.879) | 0.334 (1.167) | 0.275 (0.756) | |
| 6742 | 0.452 (0.682) | 0.512 (0.894) | 0.294 (1.230) | 0.276 (0.756) | |
| 13483 | 0.449 (0.681) | 0.515 (0.890) | 0.293 (1.229) | 0.275 (0.756) | |
| 20225 | 0.451 (0.683) | 0.517 (0.883) | 0.311 (1.186) | 0.275 (0.756) |
表5
G2PSE模型各子模型在Pepper203数据集PHT性状上的泛化能力对比"
| 模型 Model | 验证集Validation set | 测试集Test set | 差值绝对值Absolute difference | |||
|---|---|---|---|---|---|---|
| PCC | MAE | PCC | MAE | PCC | MAE | |
| AFSE | 0.639 | 0.566 | 0.520 | 0.580 | 0.119 | 0.014 |
| LFSE | 0.735 | 0.502 | 0.645 | 0.563 | 0.090 | 0.061 |
| LFESE | 0.693 | 0.544 | 0.734 | 0.517 | 0.041 | 0.027 |
| AFESE | 0.659 | 0.553 | -0.023 | 0.690 | 0.682 | 0.137 |
| HFESEⅠ | 0.669 | 0.553 | 0.415 | 0.745 | 0.254 | 0.192 |
| HFESEⅡ | 0.659 | 0.554 | 0.527 | 0.625 | 0.132 | 0.071 |
表6
G2PSE模型各子模型在Pepper203数据集FT性状上的泛化能力对比"
| 模型 Model | 验证集Validation set | 测试集Test set | 差值绝对值Absolute difference | |||
|---|---|---|---|---|---|---|
| PCC | MAE | PCC | MAE | PCC | MAE | |
| AFSE | 0.785 | 0.468 | 0.705 | 0.508 | 0.080 | 0.040 |
| LFSE | 0.806 | 0.444 | 0.756 | 0.501 | 0.050 | 0.057 |
| LFESE | 0.753 | 0.502 | 0.718 | 0.561 | 0.035 | 0.059 |
| AFESE | 0.769 | 0.467 | 0.434 | 0.701 | 0.335 | 0.234 |
| HFESEⅠ | 0.736 | 0.564 | 0.657 | 0.624 | 0.079 | 0.060 |
| HFESEⅡ | 0.768 | 0.469 | 0.712 | 0.521 | 0.056 | 0.052 |
| [1] |
李棉燕, 王立贤, 赵福平. 机器学习在动物基因组选择中的研究进展. 中国农业科学, 2023, 56(18): 3682-3692. doi: 10.3864/j.issn.0578-1752.2023.18.015.
|
|
|
|
| [2] |
doi: 10.3168/jds.2007-0980 pmid: 18946147 |
| [3] |
|
| [4] |
|
| [5] |
doi: 10.1038/s41437-022-00539-9 pmid: 35508540 |
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
doi: 10.3168/jds.2020-19789 pmid: 33663836 |
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
doi: 10.1038/s41598-024-57234-4 pmid: 38493207 |
| [15] |
|
| [16] |
doi: 10.1007/s10994-019-05848-5 pmid: 32174648 |
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
周铂涵, 梅步俊, 吕琦, 王志英, 苏蕊. 机器学习及其在动物遗传育种中的应用研究进展. 中国畜牧兽医, 2024, 51(12): 5348-5358.
doi: 10.16431/j.cnki.1671-7236.2024.12.022 |
|
|
|
| [22] |
doi: 10.1007/s00425-018-2976-9 pmid: 30101399 |
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
林泳恩, 孟越, 杜懿, 王大洋, 王大刚. 堆叠集成模型径流预报效果的影响因素研究. 水文, 2023, 43(1): 57-61.
|
|
|
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
冯盼峰, 温永仙. 基于随机森林算法的两阶段变量选择研究. 系统科学与数学, 2018, 38(1): 119-130.
doi: 10.12341/jssms13325 |
|
doi: 10.12341/jssms13325 |
|
| [36] |
孙嘉利, 吴清太, 温阳俊, 张瑾. 基于FASTmrEMMA、最小角回归和随机森林的全基因组选择新算法. 南京农业大学学报, 2021, 44(2): 366-372.
|
|
|
|
| [37] |
|
| [38] |
pmid: 16219924 |
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
李娟, 章明清, 许文江, 孔庆波, 姚宝全. 提高三元肥效模型建模成功率的主成分回归技术研究. 土壤学报, 2018, 55(2): 467-478.
|
|
|
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
doi: 10.1007/s00122-017-2988-z pmid: 28986680 |
| [1] | 梅广源, 李荣, 梅新, 陈日强, 樊意广, 程金鹏, 冯子恒, 陶婷, 赵倩, 赵培钦, 杨小冬. 基于VSURF-CA的小麦条锈病高光谱病情指数估测模型[J]. 中国农业科学, 2024, 57(3): 484-499. |
| [2] | 范鹏, 杨天乐, 朱少龙, 王志杰, 张明月, 魏海燕, 刘国栋. 长三角地区半糯粳稻外观品质的评价[J]. 中国农业科学, 2024, 57(16): 3105-3115. |
| [3] | 曹珂, 陈昌文, 杨选文, 别航灵, 王力荣. 桃果实单果重及可溶性固形物含量的全基因组选择分析[J]. 中国农业科学, 2023, 56(5): 951-963. |
| [4] | 赵静,李志铭,鲁力群,贾鹏,杨焕波,兰玉彬. 基于无人机多光谱遥感图像的玉米田间杂草识别[J]. 中国农业科学, 2020, 53(8): 1545-1555. |
|
||